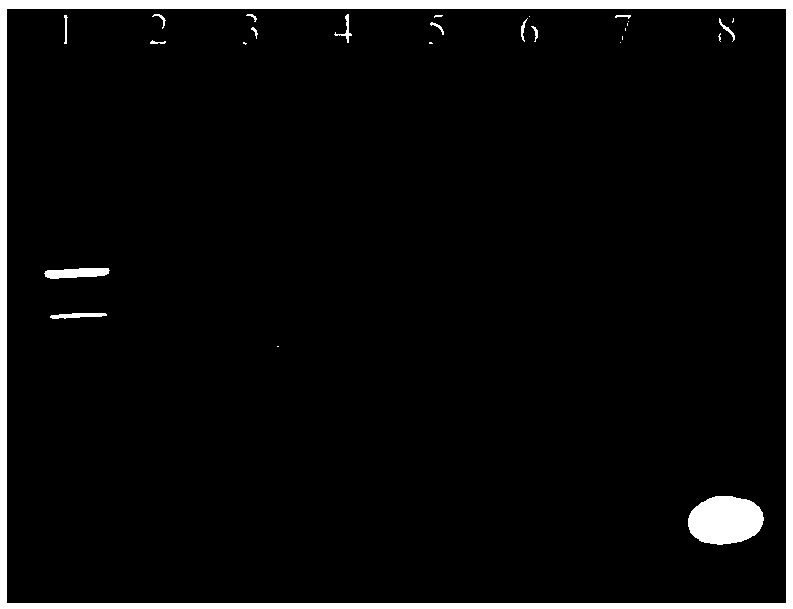PEG modified nucleic acid aptamer as well as preparation method and application thereof
A nucleic acid aptamer and reaction technology, applied in biochemical equipment and methods, medical preparations containing active ingredients, pharmaceutical formulations, etc., to achieve good stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] 1) Combine PEG5000, 0.400M 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (also known as EDC·HCl) and 0.100M N-hydroxysuccinimide The (NHS) molar ratio was 4:4:1 (50 μL each) after mixing, and shaking at 80 rpm for 15 min.
[0041] 2) Add FT6 aptamer with a volume ratio of 1:30 to PEG to 50μL of 2-(N-morpholine)ethanesulfonic acid buffer (also known as MES buffer) with a pH of 6 and then add it to Step 1) The prepared mixture was shaken at 80 rpm for 4 hours, and gel electrophoresis was performed after the reaction was completed.
Embodiment 2
[0043] 1) Combine PEG5000, 0.400M 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (also known as EDC·HCl) and 0.100M N-hydroxysuccinimide The (NHS) molar ratio is 5:5:1 (50 μL each) after mixing, and the reaction is shaken at 80 rpm for 10 min.
[0044] 2) Add FT7 aptamer with a volume ratio of 1:10 to PEG to 50μL of 2-(N-morpholine) ethanesulfonic acid buffer (also known as MES buffer) with a concentration of 20ml / L and a pH of 4 Solution), then add to the mixed solution prepared in step 1), shake for 2h at a rotation speed of 60 rpm, and perform gel electrophoresis detection after the reaction.
Embodiment 3
[0046] A PEG-modified nucleic acid aptamer preparation method, the steps are as follows:
[0047] 1) Combine PEG5000, 0.400M 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (also known as EDC·HCl) and 0.100M N-hydroxysuccinimide The (NHS) molar ratio is 2:2:1 (50 μL each) after mixing, and shaking at 100 rpm for 20 min.
[0048] 2) Add FT8 aptamer with a volume ratio of 1:50 to PEG to 50μL of 2-(N-morpholine)ethanesulfonic acid buffer (also known as MES buffer) with a pH of 7 and then add it to Step 1) The prepared mixture was shaken at 80 rpm for 5 hours, and gel electrophoresis detection was performed after the reaction.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com