Establishment and application of novel HIV-1 drug resistance detection method
A technology for HIV-1 and drug resistance, applied in biochemical equipment and methods, microbial measurement/testing, DNA/RNA fragments, etc., can solve the problems of large amount of purification reagents, large amount of nucleic acid usage, waste, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0112] Embodiment 1, the primer combination that is used for HIV-1 drug resistance detection
[0113] Primer combination I: primer DR1-1-CNB, primer DR2-1-CNBC, primer DR1-2-CNB, primer DR2-2-CNBC, primer pol-1e, primer pol-x, primer IN1 respectively in Table 1 , Primer IN1-07BC, Primer IN2 and Primer IN2-07BC were configured into primer solutions of equal concentration using denucleic acid water, and then mixed in equal volumes to obtain primer combination I (the concentration of each primer in primer combination I was the same).
[0114] Primer Combination II: Prepare the primer DR-3, primer DR1-4-CNB and primer DR2-4-CNBC in Table 1 respectively to a primer solution of equal concentration using de-nucleic acid water, and then take 2 parts by volume of primer DR-3, 1 part by volume of primer DR1-4-CNB and 1 part by volume of primer DR2-4-CNBC are mixed to obtain primer combination II (concentrations of primer DR-3, primer DR1-4-CNB and primer DR2-4-CNBC in primer combination...
Embodiment 2
[0116] Embodiment 2, establishment of novel HIV-1 drug resistance detection method
[0117] 1. Extract the total RNA from the isolated plasma sample of the patient to be tested.
[0118] 2. Using the total RNA obtained in step 1 as a template, carry out the synthesis of cDNA and the first round of nested PCR amplification. The reaction system is shown in Table 2, and the reaction procedure is shown in Table 3.
[0119] Table 2 cDNA synthesis and the first round of nested PCR amplification system
[0120] composition
Volume (μL)
PrimeScript 1Step Enzyme Mix
1
2×1Step Buffer
12.5
Primer combination Ⅰ (the concentration of each primer in the system is 2μM)
1
RNase Free dH 2 o
5.5~0.5
5~10
total
25
[0121] Table 3 cDNA synthesis and the first round of nested PCR amplification reaction program
[0122]
[0123] 3. Carry out step (1) or step (2) according to the medication situation of the pati...
Embodiment 3
[0144] Embodiment 3, specificity
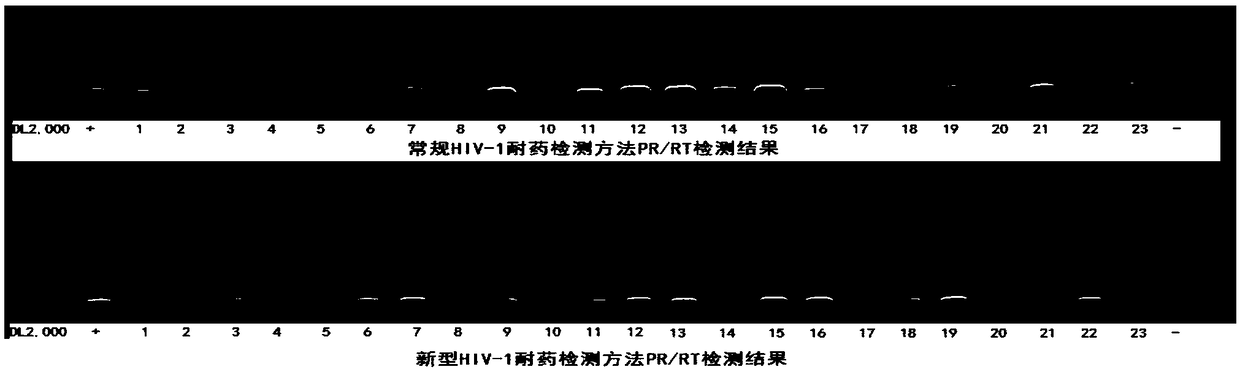
[0145] Five HTLV-infected patients with pre-clinical diagnosis and positive nucleic acid test (volunteers with informed consent; numbered HTLV25, HTLV26, HTLV36, HTLV37, HTLV38), and five HBV-infected patients with clinical diagnosis and positive nucleic acid test (informed consent) were used. Volunteers; numbered HBV-2, HBV-5, HBV-6, HBV-10, HBV-12), eight HCV-infected patients who were clinically confirmed and positive in nucleic acid tests (informed consent volunteers; numbered respectively HCV-5, HCV-70, HCV-71, HCV-75, HCV-76, HCV-93, HCV-7, HCV-56; where HCV-70 is the co-infection of HIV-1 and HCV infection) as The isolated plasma samples were used as experimental samples for specific detection.
[0146] The detection was carried out according to the method in Example 2. After cDNA synthesis and the first round of PCR amplification with primer combination I, the target gene was amplified again with primer combination II and primer combin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com