System for detecting cancer recurrence by using low-depth whole genome sequencing
A whole genome sequencing and cancer technology, applied in the field of systems for detecting cancer recurrence, can solve problems such as difficulty, high detection cost, and inability to detect false negative results, and achieve the effect of eliminating errors and low sequencing costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
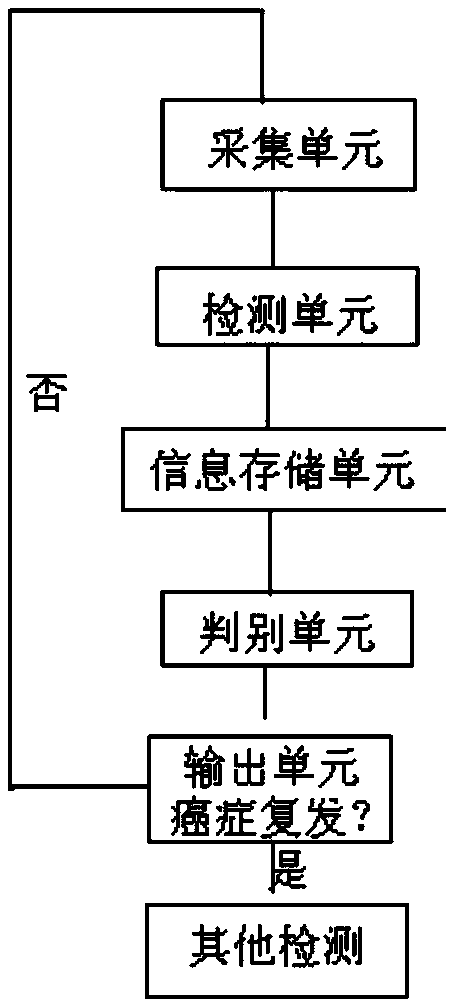
[0038] like figure 1 The system for detecting cancer recurrence using low-depth whole-genome sequencing is composed of an acquisition unit, a detection unit, an information storage unit, a discrimination unit and an output unit connected in sequence.
[0039] When in use, about 10 mL of peripheral blood from cancer patients is collected by the collection unit. The QIAGEN circulating DNA kit kit of the acquisition unit extracts 2-1000ng of ctDNA, and delivers 2-10ng to the detection unit. The WGA kit of the detection unit (such as ThruPLEX Plasma-Seq Kit, Rubicon Genomics) builds a library for ctDNA sequencing, and performs low-depth whole-genome sequencing on the DNA of the built library on a high-throughput NGS sequencer such as Illumina NextSeq, with an average sequencing depth of 1x Or 0.5x, and send the produced sequencing data to the CNV analysis unit in BAM file format for analysis. The analysis software of the CNV analysis unit can be Nexus Copy Number or CNVKit, etc.,...
Embodiment 2
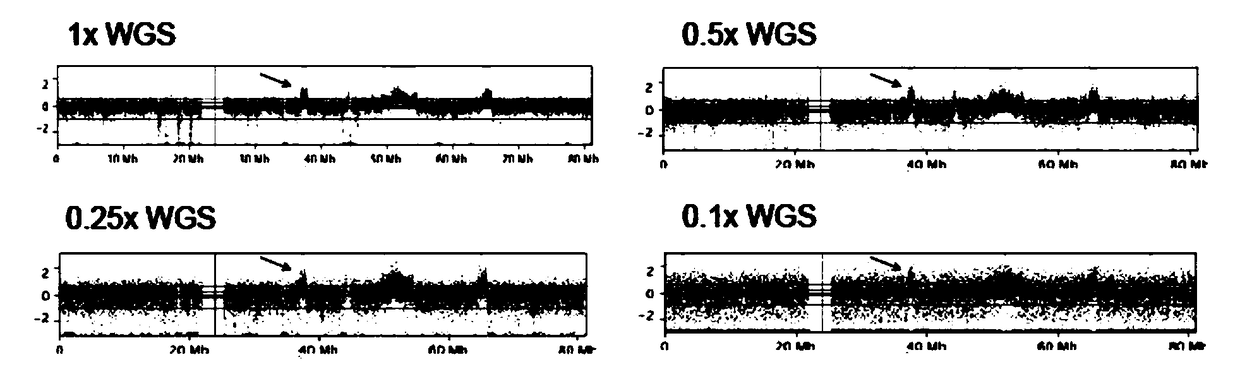
[0040] Example 2 Effect of Different Sequencing Depths on ctDNA CNV Results of Breast Cancer
[0041] The other operations are exactly the same. The sequencing depth of the detection unit is set to 1x, 0.5x, 0.25x and 0.1x respectively, and the same breast cancer patient is sampled for detection. The sequencing data is as follows: figure 2 shown. The arrow is the CNV repeat of the chr17q12 fragment containing the ERBB2 gene. It can be seen from the figure that the CNV data quality of 1x or 0.5x sequencing depth is good, the data quality of 0.25x data begins to decline, and the data of 0.1x is basically unusable. Therefore, the detection unit of the system of the present invention sets the sequencing depth to 1x or 0.5x, which ensures low cost and high quality.
Embodiment 3
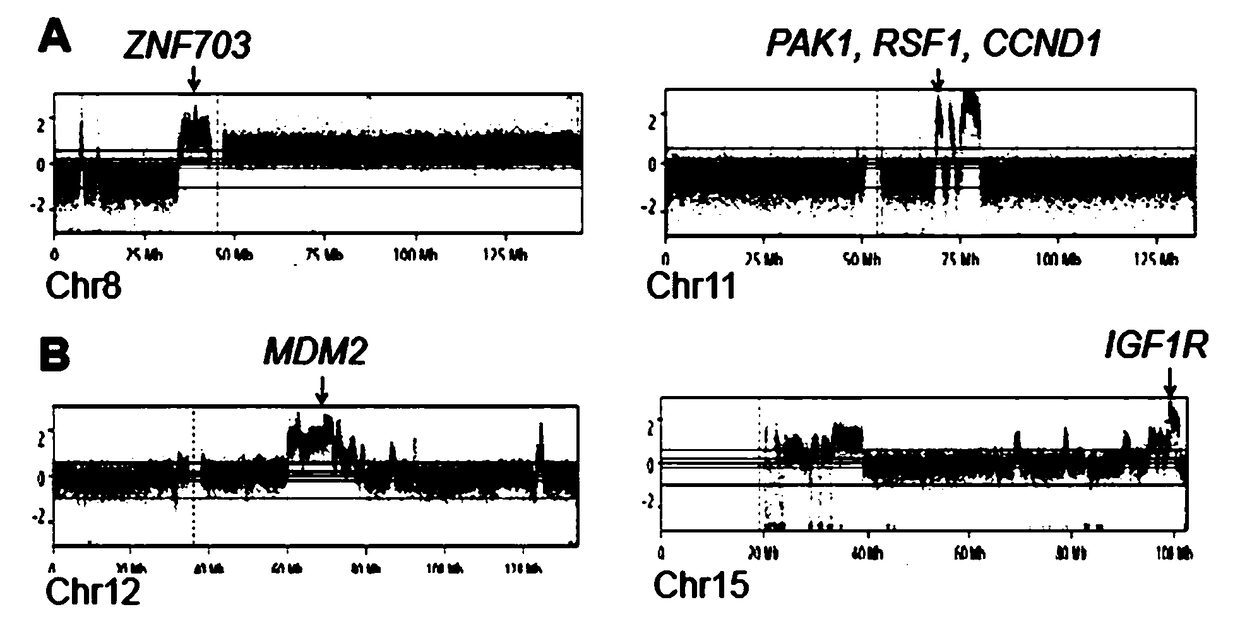
[0042] Embodiment 3 Accuracy Research
[0043] The system of Example 1 of the present invention was used to perform 0.5x low-depth whole-genome sequencing on the ctDNA of two breast cancer patients (patient A and patient B), and the CNV results detected by the detection unit were as follows: image 3 shown. At the same time, the ctDNA of the two patients was detected by qPCR method, and the results were as follows: Figure 4 shown. The results of the two methods were compared, and the gene duplications in the CNV results were confirmed in the qPCR results, showing a high degree of consistency.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com