Method for realizing genome editing and accurate and targeted knock-in of genes in fishes
A genome editing and gene knock-in technology, applied in the biological field, can solve the problems of multi-copy insertion, large workload, and low efficiency of fixed-point integration, and achieve the effect of reducing cost and improving efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1 Preparation of wild-type Cas9 or Cas9n-D10A sequence and sgRNA
[0048] The wild-type Cas9 or Cas9n-D10A sequence was added to the nuclear localization sequence (the sequence is CCGCCACC), and then the SP6 promoter sequence (the sequence was CATACGATTTAGGTGACACTATAG) was added upstream of the entire sequence, and the capping was obtained using an in vitro transcription kit (capped) and polyA-added mRNA, the obtained Cas9 or Cas9n-D10A mRNA was mixed in different combinations and frozen at -80°C for later use. The sgRNA has a T7 promoter sequence upstream and a downstream sequence partially complementary to the upstream through DNA synthesis, and double-stranded DNA is obtained by PCR amplification, which is obtained using an in vitro transcription kit.
[0049] DNA template sequence for sgRNA synthesis Primer synthesis sequence
[0050]
Embodiment 2
[0051]Example 2. Mapping potential nicks at the target site of integration of zebrafish Ndr2 and GFAP
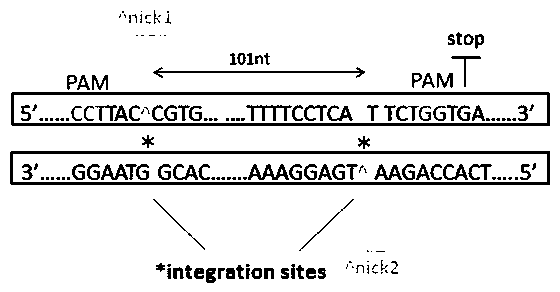
[0052] Query and download the zebrafish Ndr2 genome sequence (NM_139133) on the genome database (such as NCBI: https: / / www.ncbi.nlm.nih.gov / ), and perform PAM analysis on the sequence of the region to be integrated, according to figure 1 The spacing shown selects the two PAM sites with higher scores.
[0053] 以斑马鱼Ndr2为例,分析结果如下:agcagcaggacggggccagtctcctgcacactgccggggcctcgaagtttctgttttctagaaataagaaagaagtcaagcgaggacgggccctcaggagccgcaggggccgccg(PAM1)ggggcc(插入区域)acctgtcaggagcccagagctgcagagaacaccactgcacaagagcaccacctgcagg(PAM2)agggtggacatgcatgtggattttaaccagatcggatggggctcctggatcgtgttccctaagaagtacaa
[0054] Taking zebrafish GFAP as an example, the analysis results are as follows:
[0055] caattaagttgtccccaaaaacatctcaagaattgtgttgattctgctaattcttcattttacacattctctctttcgcagatcattaaagagtcca(PAM1)ctacggagaggaaggatctgccataa(终止子)tgaggctcagacactggcttggctgcagaagaagatctccttcactaatgcttcagg (PAM...
Embodiment 3
[0056] Example 3. Preparation of exogenous donor DNA Ndr2-linker-Dendra2 and GFAP-p2AV1-NpHR3.0-EYFP-p2AV2-hChR2-mCherry-IRES-WGA-Cre (GFAP-5.5 kb)
[0057] Such as figure 2 As shown, when preparing an exogenous donor, first, we use primers to amplify and purify the 5' homology arm (5' HA), the insert fragment, the 3' homology arm (3' HA) and the vector backbone ; Then, the 5'HA, insert and 3'HA were bridging PCR in pairs to obtain three PCR fragments connected together; finally, the 5'HA-insert-3'HA product and the vector backbone fragment were passed through In-Fusion The HD cloning kit (Clonetech, 639649) was seamlessly connected and transformed into E. coli, and the exogenous donor DNA was obtained after identification by picking clone sequencing, and Ndr2-linker-Dendra2 and GFAP-p2AV1-NpHR3.0-EYFP- p2AV2-hChR2-mCherry-IRES-WGA-Cre (GFAP-5.5 kb).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com