A polypeptide fragment specifically binding to the serum of Babesia patients and its application
A technique for polypeptide fragments and babesiosis, which is applied in the field of biomedicine and clinical applications, can solve the problems of complex preparation process, unstable activity, and high cost, improve specificity and sensitivity, and overcome missed detection and misdiagnosis of parasites Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1 The biological characteristics of Babesia microti merozoite surface antigen BMSA
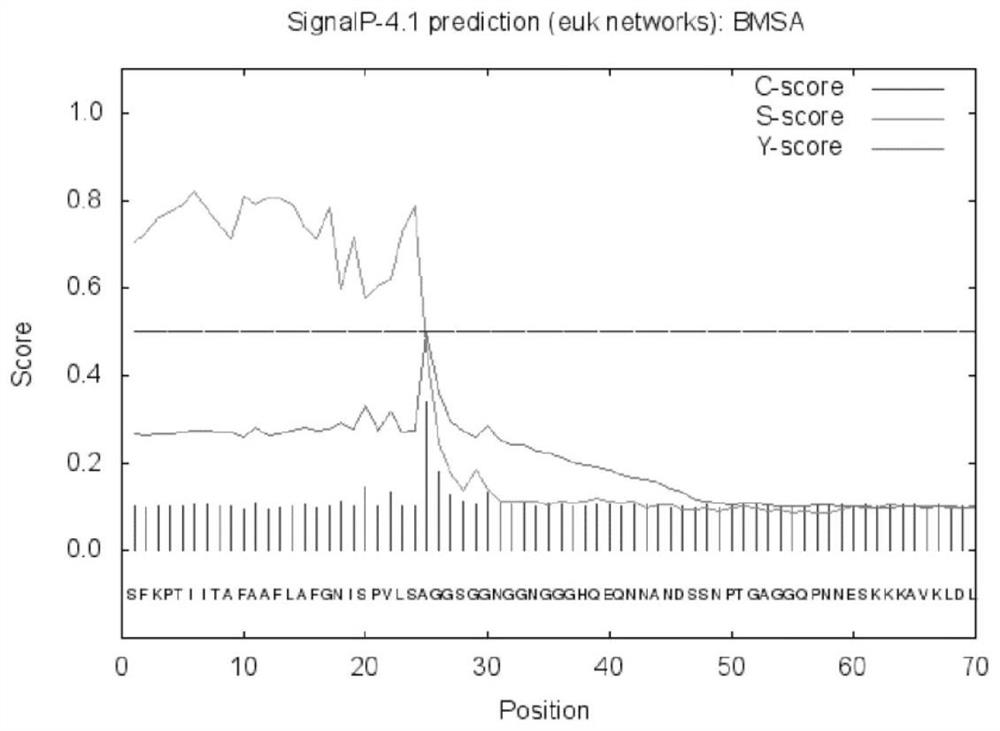
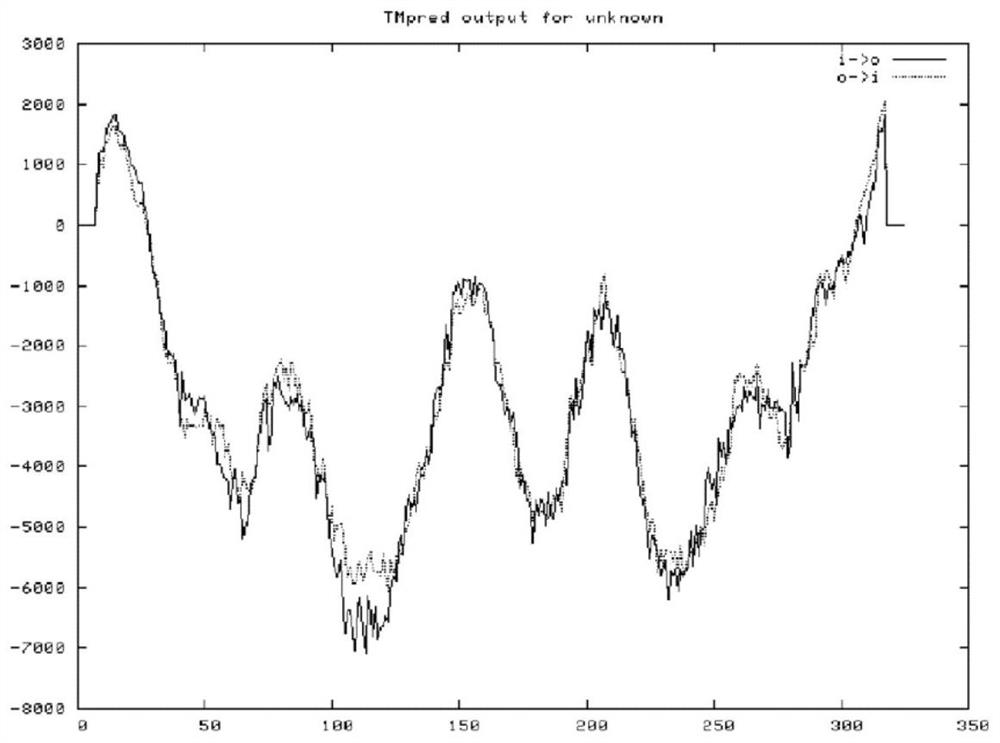
[0022] According to the Babesia microti merozoite surface antigen BMSA gene sequence (such as figure 1 Shown) The deduced protein is composed of 326 amino acids (as shown in Sequence 2), and the size is 35.117kDa; the isoelectric point pI is predicted to be 5.21 by ExPAS software, and the 302nd amino acid is predicted by the PredGPI software to be the GPI anchor site, SignalP4 .1 The software predicts that the N-terminal 24 amino acids are signal peptides (such as figure 2 shown); TMPred software predicts the transmembrane segment from extracellular to intracellular, from amino acid 310 to 326 (Score2066) (eg image 3 shown), the PotParam software predicts that negatively charged amino acids account for 53%, and the proportion of positively charged amino acids is 43%; homology modeling predicts the three-dimensional structure of BMSA as Figure 4 shown.
Embodiment 2
[0023] Example 2 Epitope Analysis and Verification of the Babesia vole merozoite surface antigen BMSA
[0024] According to the Disco Tope 2.0 software, there are 6 antigenic epitopes (as shown in Table 1) for the B-cell epitope of the Babesia vole merozoite surface antigen BMSA, and the peptide synthesis of these 6 epitopes is carried out by artificial peptide synthesis. Synthesize; then screen and identify the epitope by indirect ELISA method through the serum of mice infected with Babesia, the method is as follows: Add 100 μL / well Coating Buffer (0.5 μg of artificially synthesized BMSA protein polypeptide) to the 96-well microtiter plate , in a wet box, coated overnight at 4°C; remove the coating solution, wash the plate with PBST, add 1% skimmed milk-PBS at room temperature, and block for 1 hour; add double-diluted Babesia-infected mouse serum and BMSA After immunization, mouse serum was 100 μL / well (1:3200 1:6400, PBS doubling dilution), incubated at room temperature for ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com