Construction method, construction kit and application of optic atrophy animal model
A technology of optic atrophy and animal models, applied in biochemical equipment and methods, animal/human peptides, chemical instruments and methods, etc., can solve the problems of lack of animal models of optic atrophy, achieve rich categories, short modeling time, and promote The effect of research and development
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0045] In this embodiment, mice are taken as an example to illustrate the method for constructing the mouse model of optic atrophy in this embodiment. The specific operation is as follows:
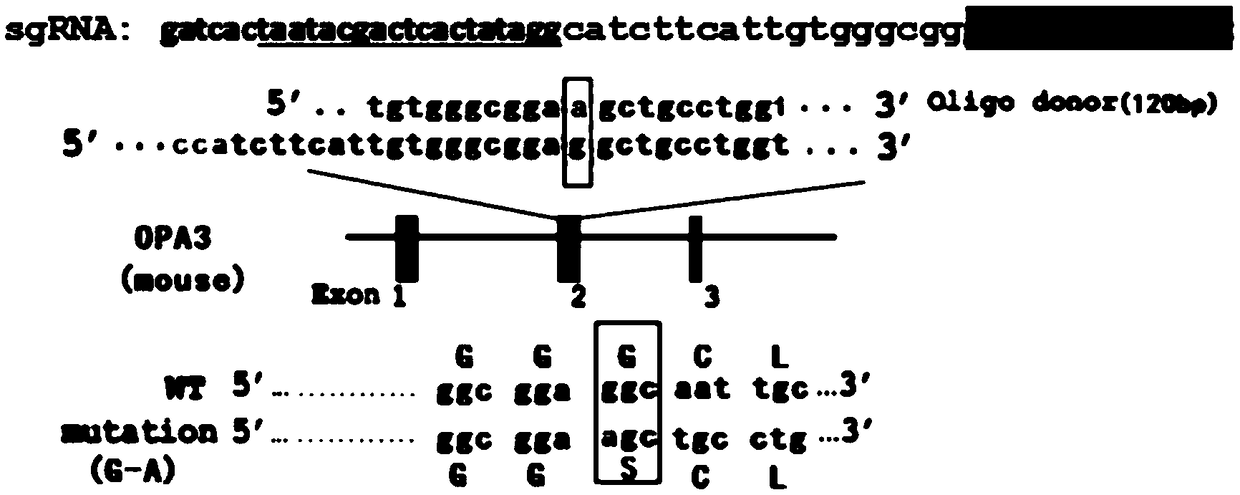
[0046] (1) Using the CRISPR design tool (online design tool http: / / tools.genome-engineering.org), select the CRISPR / Cas system knockout target, that is, the 18bp DNA ending with NGG as the target sequence: CATCTTCATTGTGGG CGG (5'-3'); the underline is the position of the PAM motif.
[0047] (2) For the above-mentioned target, an oligonucleotide chain with a T7 promoter sequence and a 60bp target sequence (ie, the sgRNA precursor sequence) synthesized in vitro, the sequence of the sgRNA precursor sequence is as follows:
[0048] GATCACTAATACGACTCACTATAGG CATCTTCATTGTGGGCGG GTTTTAGAGCTAGAAAT; target sequence is underlined.
[0049] (a) Using this as a template, perform PCR reaction with high-fidelity KOD enzyme to obtain a DNA template that can transcribe sgRNA.
[0050] PCR reaction s...
experiment example
[0074] Phenotypic Characterization of a Mouse Animal Model of Optic Atrophy
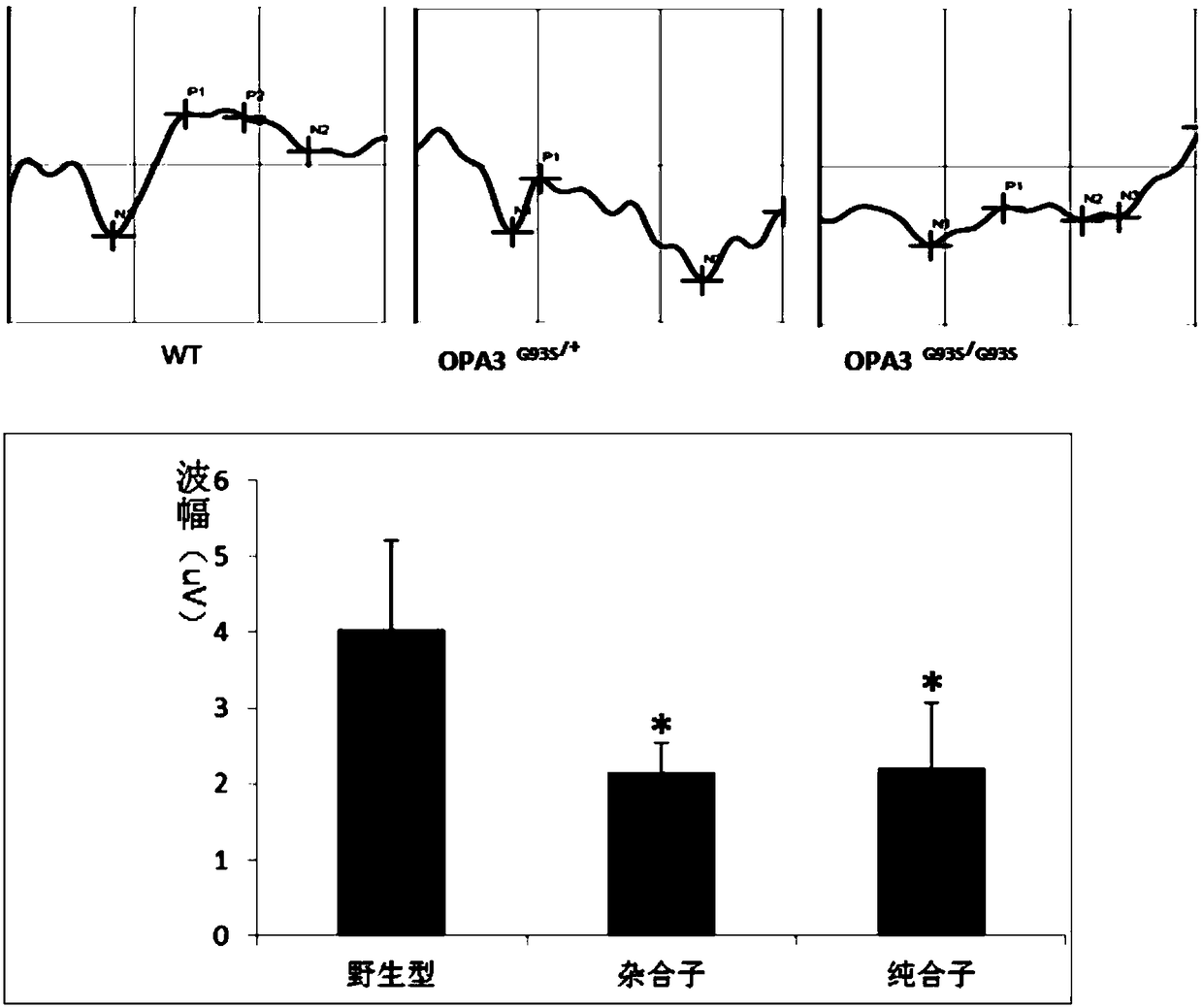
[0075] (1) Measurement of visual evoked potential (VEP)
[0076] Take the mice under the natural light adaptation state, respectively connect the recording electrode to the cheek, the reference electrode to the cranium, and the ground electrode to the tail, and use the visual electrophysiological recorder (VEP) to measure the visual evoked potential, and detect the waveform (N1 wave and P1 wave ) is abnormal.
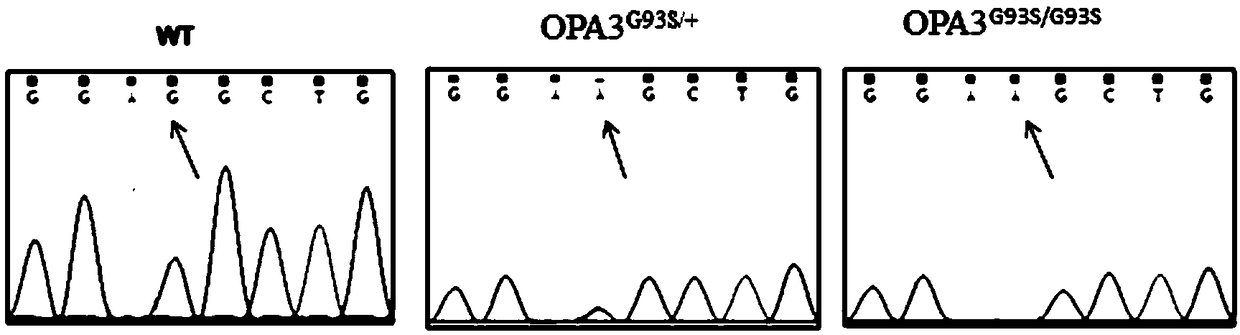
[0077] The result is as figure 2 As shown, heterozygous OPA3 G93S / + and homozygous OPA3 G93S / G93S Both animal models showed abnormal visual evoked potential (VEP), and the amplitude difference between N1 wave and P1 wave decreased.
[0078] (2) Ex vivo tissue detection: After the mice were sacrificed, the eyeball and optic nerve were bluntly separated, the scissors went deep into the deep orbit, cut off the optic nerve, and the eyeball and optic nerve were taken out.
[0079] (a) Retinal...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com