Single nucleotide polymorphism analysis method based on magnetic functionalized microfluidic chip
A single nucleotide polymorphism, microfluidic chip technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problem that the single nucleotide polymorphism analysis method has not been reported, and achieve Good biocompatibility and hydrophilicity, maintaining activity, efficient separation and detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] (1) Preparation of graphene oxide: 0.5g graphite powder and 0.5gNaNO 3 Added to 23 mL of concentrated H 2 SO 4 In the ice bath, slowly add 3gKMnO 4 , stir well to make it evenly mixed, transfer the solution to a water bath at 35°C and stir for 1 hour to form a beige paste, add 40mL of ultrapure water, stir at room temperature for 30min, add ultrapure water to dilute to 140mL, drop by drop Add 3 mL of 30% HO 2 o 2 , the solution changed from dark brown to bright yellow, and the obtained product was filtered while hot, and the product was centrifuged with ultrapure water until the supernatant was neutral, and centrifuged at 8000r / min for 2min to obtain graphene oxide (GO).
[0029] (2) Preparation of GO@Fe 3 o 4 Nanocomposite material: Dissolve 40mg GO in 20mL ultrapure water, ultrasonically disperse for 3h, and then pass N 2 Under the condition of heating to 70 ℃ and keeping it for 30min, then add 21mg FeCl 3 ·6H 2 O and 72 mg FeCl 2 4H 2 O, stirred for 40min;...
Embodiment 2
[0033] Preparation of GO@Fe 3 o 4 Functionalized PDMS microfluidic chip separation channel: first rinse the PDMS microfluidic separation channel with ultrapure water for 5 minutes, then place a permanent magnet with a length of 2 cm on the top and bottom of the chip, and vacuum the GO prepared in Example 1 @Fe 3 o 4 The solution is pumped into the separation channel, GO@Fe 3 o 4 It was immobilized in the PDMS microfluidic separation channel, and the modified microfluidic chip was placed for 1 hour, and the separation channel was continuously washed with running buffer solution for 5 minutes to form GO@Fe 3 o 4 Functionalized PDMS microfluidic chip separation channel.
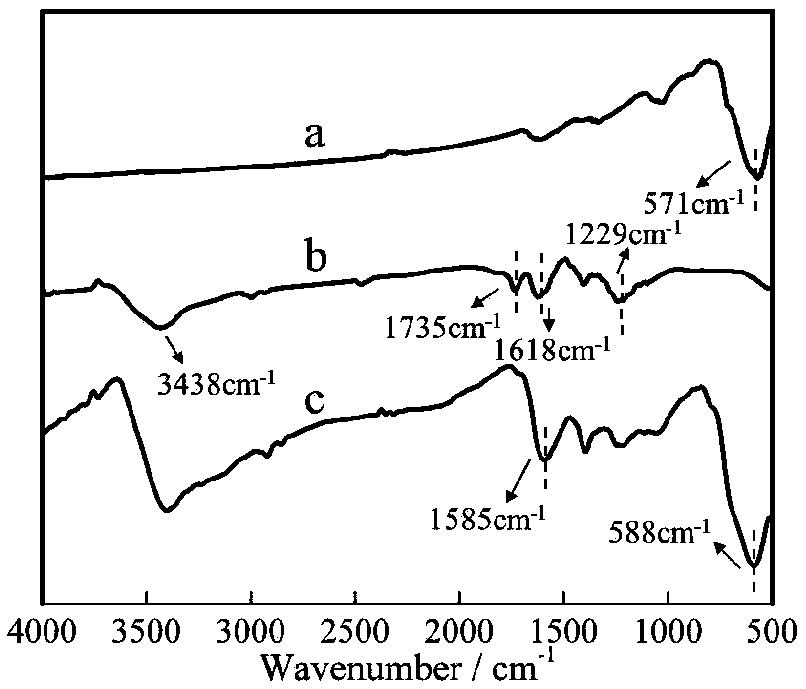
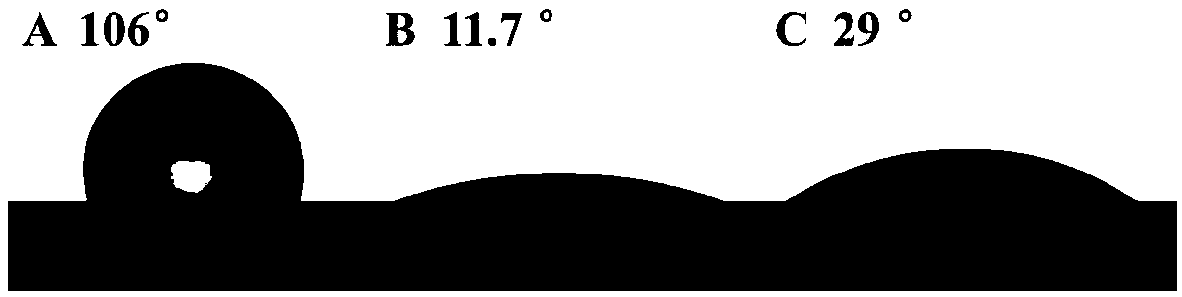
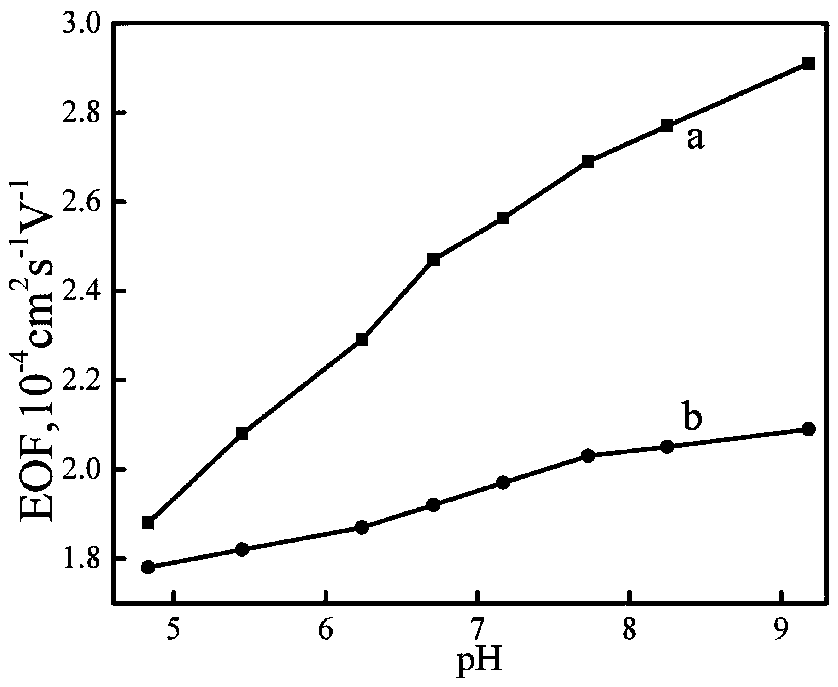
[0034] GO@Fe will be produced 3 o 4 The functionalized PDMS microfluidic chip was used to characterize the contact angle, and the PDMS bare chip and GO-modified PDMS chip were used as controls. The results are as follows: image 3 shown. Depend on image 3 A shows that the contact angle of the PDMS mi...
Embodiment 3
[0037] GO@Fe 3 o 4 Analysis of Single Nucleotide Polymorphism by Functionalized PDMS Microfluidic Chip
[0038]Design a methylene blue-labeled probe DNA (P1, 5'-methylene blue-TCAACATCAGTCTGATAAGCTA-3') that is completely complementary to mircoRNA-21 (T1, 5′-UAGCUUAUCAGACUGAUGUUGA-3′), P1 hybridizes with T1 to form a completely complementary double strand (P1T1), P1 hybridizes with the single-base mismatch strand sm-mircoRNA-21 (M1, 5′-UAGCUUAUAAGACUGAUGUUGA-3′) of T1 to form a single-base mismatch double strand (P1M1). Mix different concentrations of T1, 50 μL of 2.5 μM P1 and 20 μL of hybridization buffer solution (20 mM Tris-HCl buffer solution, pH 7.4), add ultrapure water to make the final volume of the solution 200 μL, and perform hybridization reaction at 37°C for 2.5 h. The hybridization reaction between M1 and P1 was carried out in the same steps as above. The hybridization reaction between different concentrations of T1 and different concentrations of M1 and 0.5 μ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com