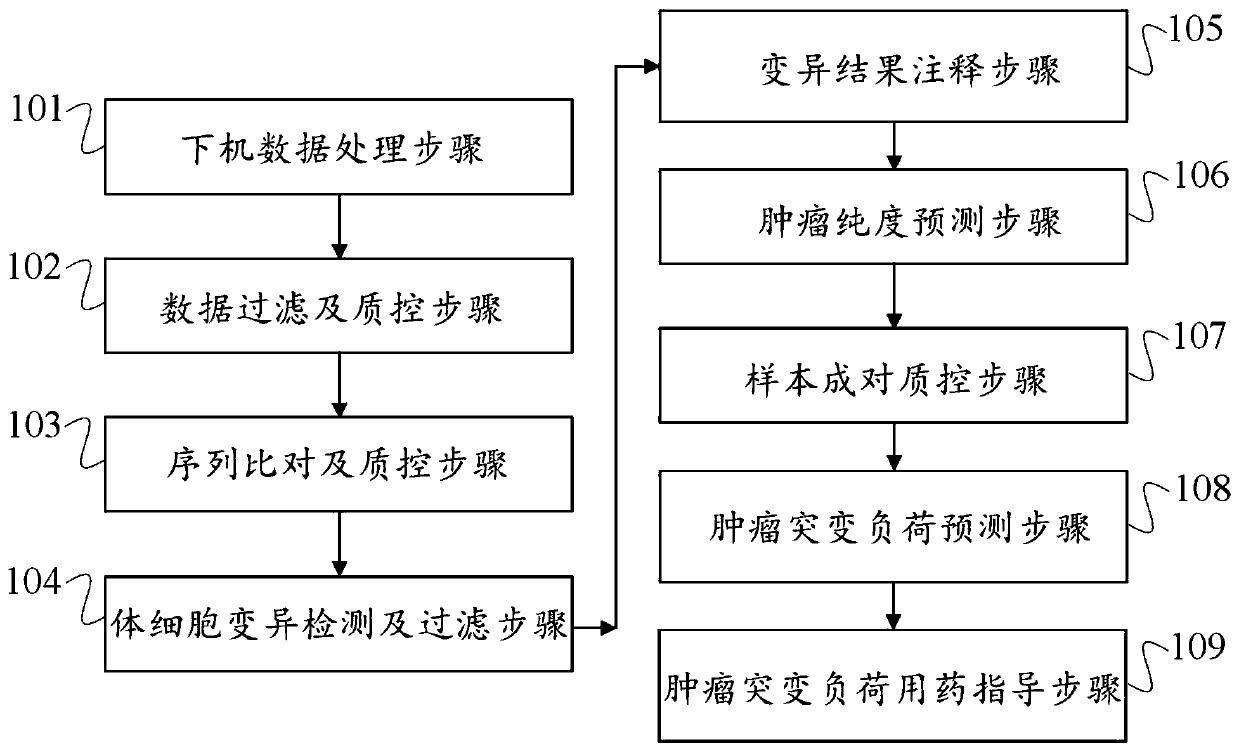
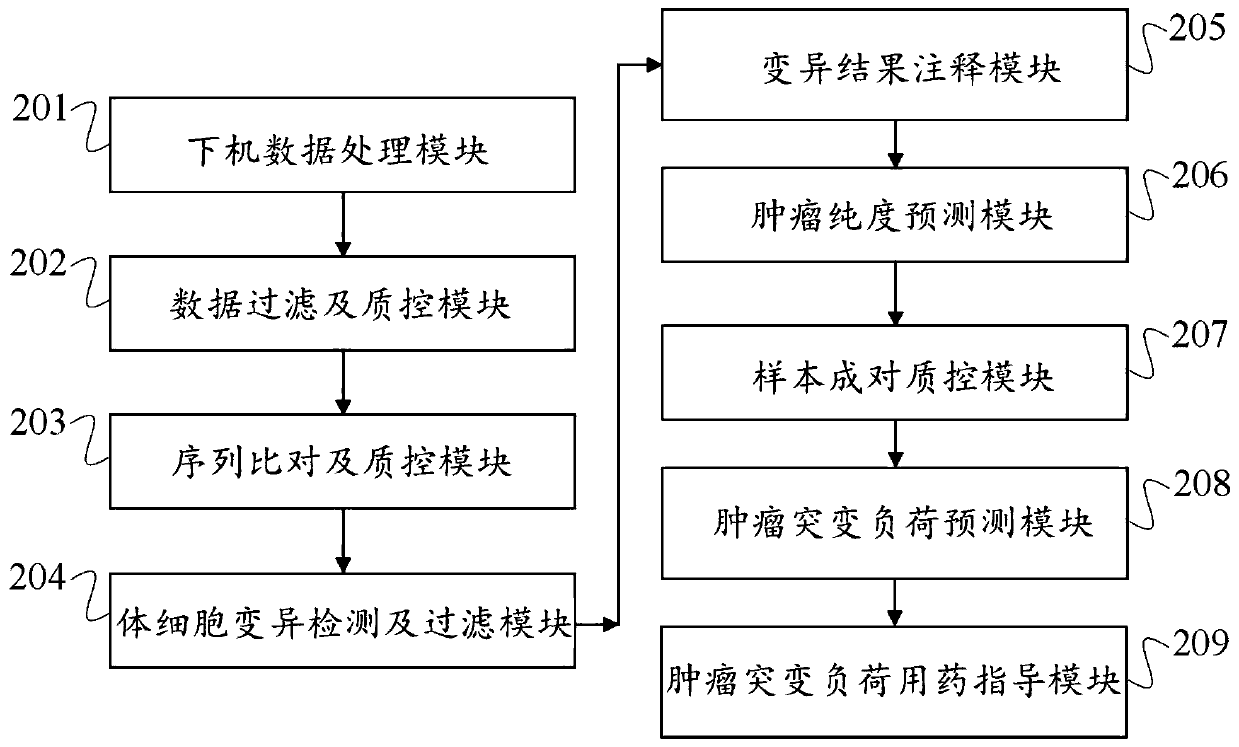
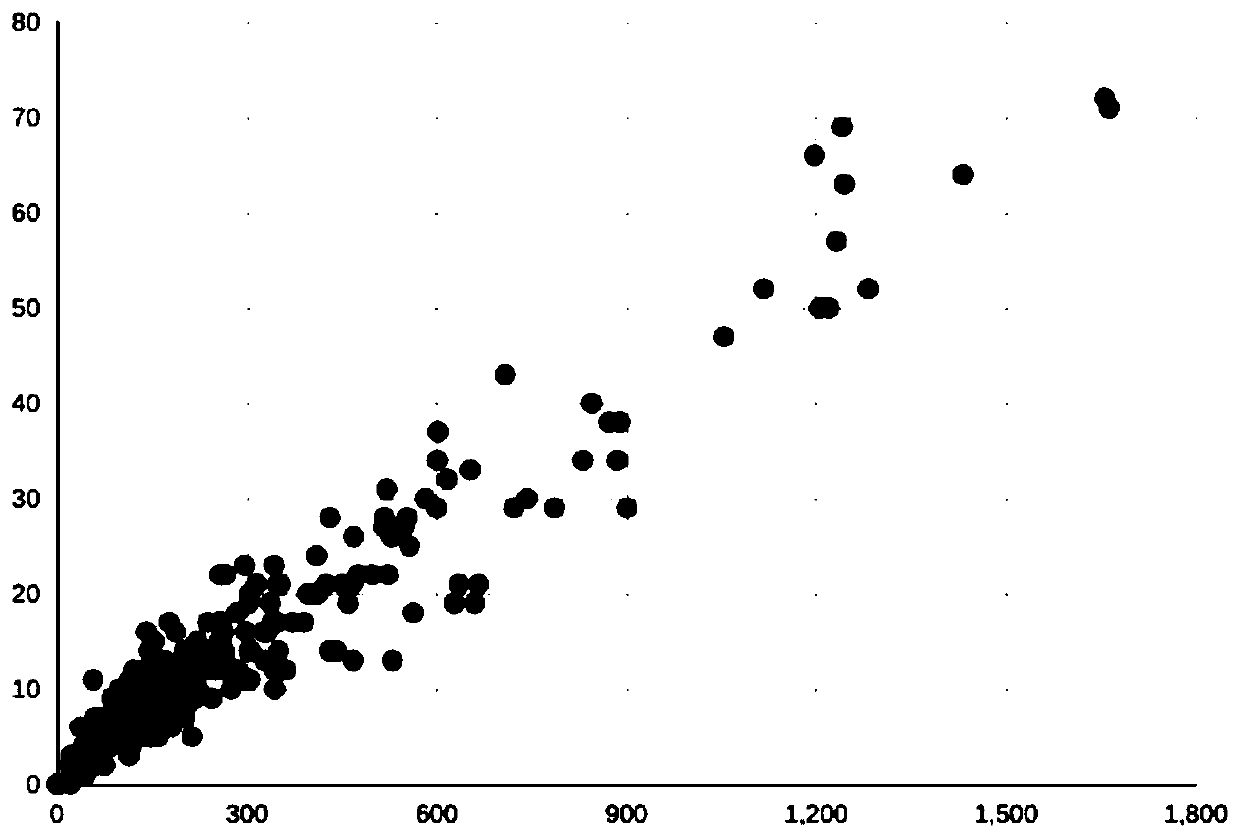
A method, device and storage medium for detecting tumor mutation load
A technology of mutation load and detection method, applied in the directions of proteomics, genomics, instruments, etc., can solve the problems of long cycle, inability to effectively distinguish somatic mutation from germ cell mutation, and unsuitable for clinical application, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0165] In order to ensure that the capture area of the gene chip can truly and accurately reflect the changing trend of tumor mutation load on the whole human genome, this application specifically provides a preparation method of the gene chip, including the design of the capture area of the chip, which specifically includes the following steps:
[0166] The statistical steps of exon mutation probability include:
[0167] 1) Count the number of mutated bases on each exon of each gene in the COSMIC database, and divide the number of mutated bases on the exon by the total length of the corresponding exon to obtain the occurrence of the exon The probability of mutating bases, marked as pa;
[0168] Among them, COSMIC database reference S.A.Forbes et al., "COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer," Nucleic Acids Res., vol.43, no.D1, pp.D805–D811, Oct.2015.
[0169] 2) In the ICGC database, the number of mutated bases on each exon of each ge...
Embodiment 1
[0185] Traditional tumor mutation burden detection usually uses whole exome sequencing, which has the disadvantages of high cost and long cycle. In order to solve this problem, a targeted capture chip is designed in this example, which only captures specific gene sequences and performs sequencing, which effectively reduces the amount of sequencing data and achieves the purpose of saving costs and shortening the cycle.
[0186] The design process of the target capture chip is:
[0187] 1) According to the mutation information collected in the COSMIC database, count how many bases are mutated in each exon of each gene, divide the total number of mutations by the total length of the exon, and obtain the number of mutated bases in each exon probability.
[0188]2) According to the sample data collected by the ICGC database (https: / / icgc.org / ), count how many samples are mutated on each exon of each gene, divide the number of mutation samples by the total number of samples, and ge...
Embodiment 2
[0201] Traditional tumor genome detection methods usually use single-sample detection of tumors, and studies have found that this method cannot effectively distinguish between somatic mutations and germ cell mutations. This defect has little impact on conventional targeted detection, but has a greater impact on tumor mutation burden detection. In order to solve this problem, this example adopts the method of paired detection, and detects tumor tissue and control samples at the same time, and cooperates with subsequent analysis methods to obtain somatic mutations. Wherein, the control sample is paracancerous tissue or peripheral blood.
[0202] In this example, the sequencing sequence error correction technology is introduced in the experimental method: the current mainstream next-generation sequencing method uses the 150bp Paired End method for sequencing, and generates two 150bp reads. Due to the characteristics of the library construction method, the length of the insert fr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com