Microorganism identification method
An identification method and microbial technology, applied in biochemical equipment and methods, microbial measurement/inspection, biological material analysis, etc., can solve problems such as difficult identification of closely related species, which protein is not determined, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0123] Hereinafter, experiments performed to confirm the effects of the present invention with regard to the order of selection of the marker proteins in the present invention will be described.
[0124] (1) The strains used and the medium for culture
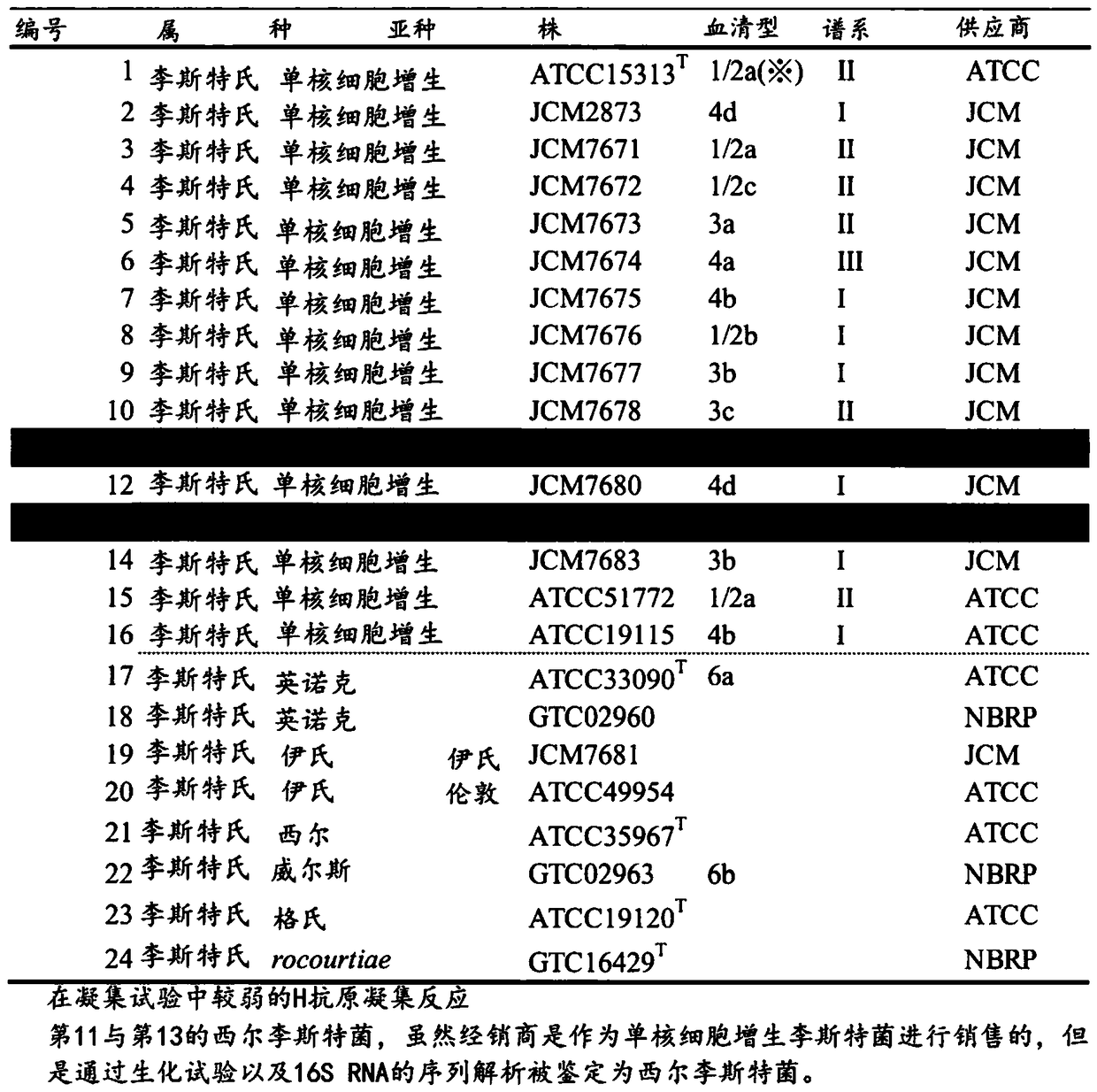
[0125] For the construction of the protein quality database, 14 strains of Listeria monocytogenes, 2 strains of Listeria innocuae, 2 strains of Listeria eziri, 2 strains of Listeria sierra, 3 strains of Listeria welsii were used bacteria, Listeria gasseri and Listeria roseri respectively, a total of 24 strains ( image 3 ). It can be obtained from the National Biological Resources Project of Japan (Japan, Gifu City, Gifu University, Department of Pathogenic Bacteria, NBPR), the American Type Culture Collection (USA, Maryland, Rockville, ATCC, American Type Culture Collection), the Japan Microorganism Collection Center (Japan, Tsukuba City, RIKEN Biological Resource Center, JCM, Japan Collection of Microorganisms) and the Biogen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com