Fluorescence in situ hybridization probe for detecting KIAA1549-BRAF fusion gene and preparation method and application thereof
A KIAA1549-BRAF, fluorescence in situ hybridization technology, applied in the field of molecular biology, can solve the problems of incomplete distinction between positive and negative samples, inability to distinguish break signal points, short distance, etc., and achieve high signal-to-noise ratio and short hybridization time Short, wide selection of effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1 Preparation of KIAA1549-BRAF fusion gene rapid detection probe
[0046] The preparation of the KIAA1549-BRAF fusion gene rapid detection probe of the present invention includes the following steps:
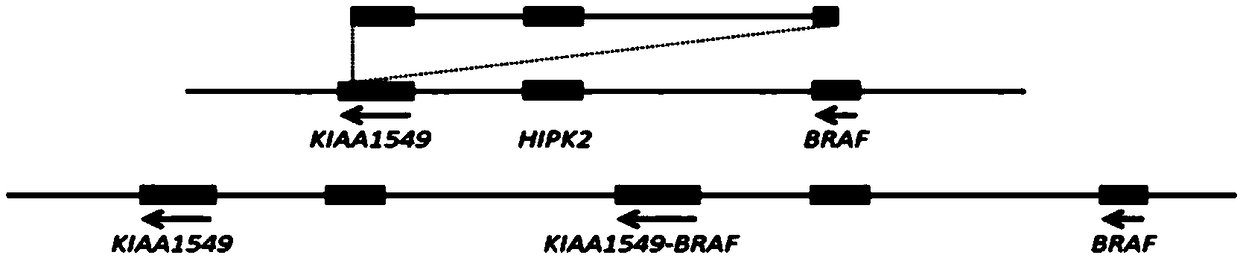
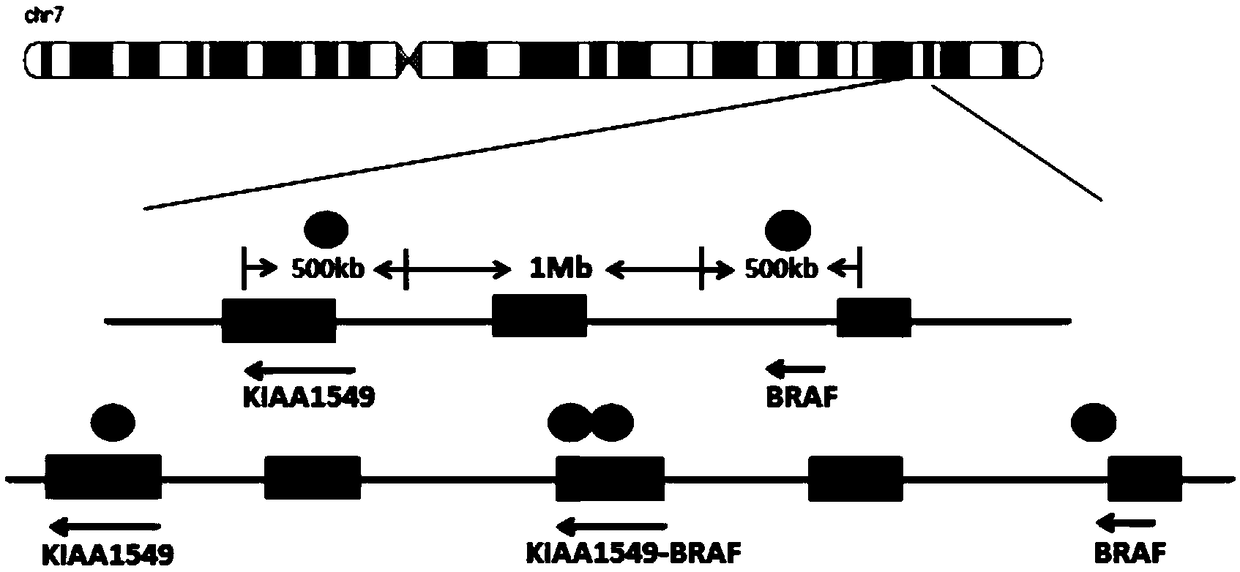
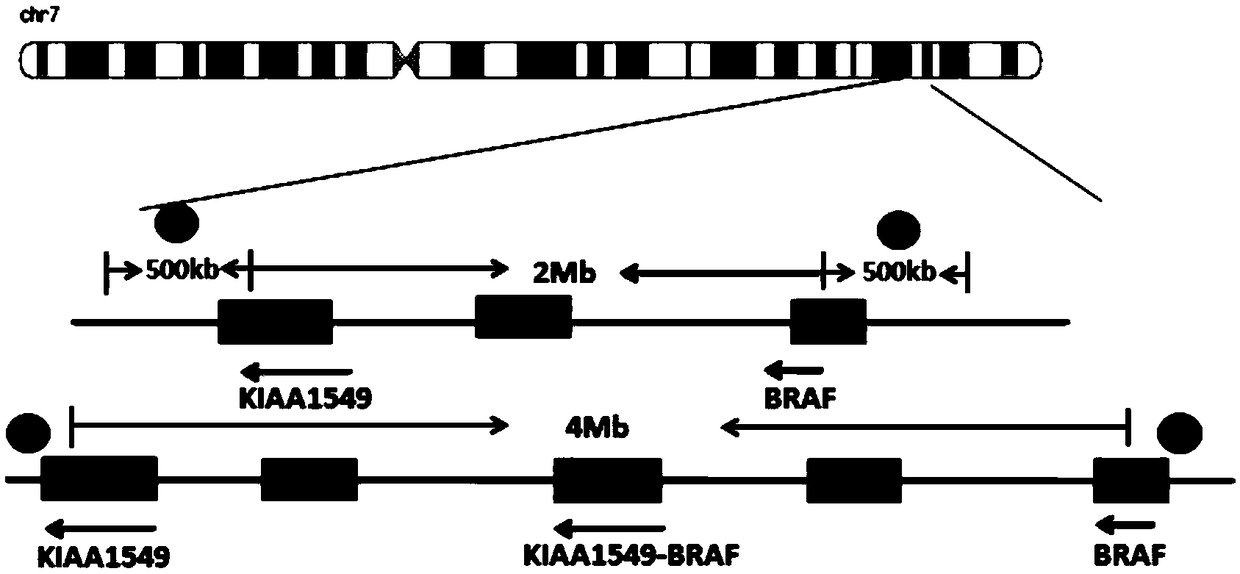
[0047] (1) Download from UCSC Genome Browser the genomic non-repetitive sequence of the region covered by the KIAA1549 and BRAF gene probes. The KIAA1549 probe covers the region: chr7 138,361,140-138,861,139 (starting position is the direction of exon 16 of KIAA1549 gene toward the centromere 500Kb region); BRAF gene probe coverage area is: chr7 140,787,547-141,287,546 (starting position is the 500kb region from exon 9 of BRAF gene to the telomere direction); the obtained sequence is saved in fasta format, and the repetitive sequence region in the genome is replaced with N;
[0048] (2) Use the perl plug-in program chunks.pl to divide the genomic non-repetitive sequence in the area covered by the KIAA1549 gene probe obtained in step (1) into 1kb blocks; import the divid...
Embodiment 2
[0055] Example 2 Method of using KIAA1549-BRAF fusion gene rapid detection probe
[0056] The method for using the KIAA1549-BRAF fusion gene rapid detection probe prepared in Example 1 to detect the KIAA1549-BRAF fusion gene specifically includes the following steps:
[0057] (1) Sample processing: Take the paraffin section sample with a thickness of 4 to 5 microns that has been baked, and immerse it in preheated 65°C dewaxing agent I for 10 to 15 minutes; take out the slide and immerse it in preheated 65°C dewaxing Reagent II for 10-15 minutes; after taking out the slide, immerse it in 100% ethanol, 85% ethanol, and 70% ethanol for 2 to 3 minutes at room temperature; take out the slide and immerse it in preheated 65℃ deionized water for 3~ 5 minutes; take out the slide and soak in 95℃ deionized water for 30-40 minutes, pre-cool the slide in cold water for 1 min; take out the slide and immerse it in preheated 37℃ protease working solution for 20-40 minutes; take out the slide and s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com