Molecular marker closely linked to salt tolerance of tomato and application thereof
A technology of salt tolerance and tomato, applied in the field of tomato breeding and molecular biology, can solve the problems of ion poisoning and lack of versatility, and achieve the effects of wide application, speeding up the breeding process, and improving the efficiency and accuracy of selection.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
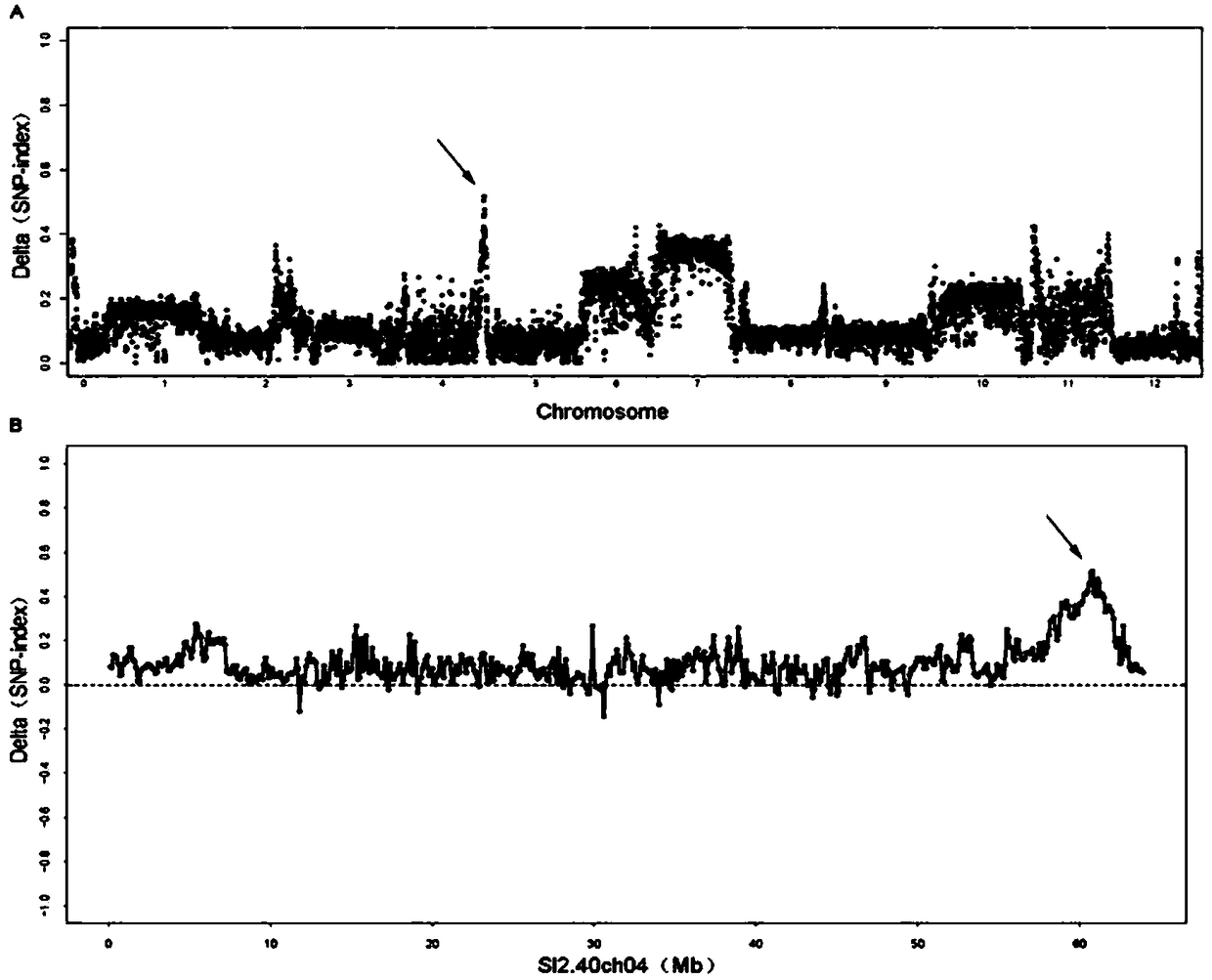
[0030] A tomato salt-tolerant molecular marker STQ4 obtained by the following method:
[0031] 1. Group building
[0032] Salt-sensitive cultivated tomato TS-3 and salt-tolerant wild gooseberry tomato TS-21 are both resequenced tomato germplasm resources (Linet al.Nature Genetics2014,46:1220-1226), with TS-3 as the female parent, TS -21 is the male parent crossed to obtain F1, and then self-crossed to obtain F2, and the RIL population was constructed by the method of single son generation.
[0033] 2. Identification of salt tolerance
[0034] (1) The RIL6 population was planted in a 7×7cm black square pot containing the same amount of nutrient soil, and there were 2 replicates. The plants were grown in a greenhouse with a temperature of 25±2°C, a photoperiod of 16h light / 8h dark, a light intensity of 3500Lux, and normal fertilizer and water management.
[0035] (2) When the seedlings have 2 leaves and 1 heart, take a small amount of samples of each line of plants and freeze...
Embodiment 2
[0044] Verification of tomato salt-tolerant molecular marker STQ4:
[0045] Using the obtained salt-tolerant molecular marker primers, 30 parts of salt-sensitive materials and 30 parts of salt-tolerant materials screened in step 2 of Example 1 were used for salt-tolerance identification. The steps are as follows:
[0046] (1) Using tomato genomic DNA as a template, the concentration is 80-120ng / μl; molecular marker STQ4 is used for PCR amplification, the sequence of the forward primer is SEQ ID NO.1, and the sequence of the reverse primer is SEQ ID NO.2. PCR reaction system: the total volume is 15 μl, and the specific components are as follows: ddH 2 O 10.4μl, 10×PCR Buffer1.5μl, 1.2μldNTPs (2mmol / L), 0.08μl Taq DNA polymerase (5U / μl), 0.4μl forward primer (10ng / μl) (sequence SEQ ID NO.1), 0.4 μl reverse primer (10 ng / μl) (sequence SEQ ID NO.2) and 1.0 μl DNA template (100 ng / μl).
[0047] (2) The PCR reaction was carried out on the S1000 PCR machine produced by Bio-Rad Comp...
Embodiment 3
[0052] Application of tomato salt-tolerant molecular marker STQ4 in tomato salt-tolerant breeding:
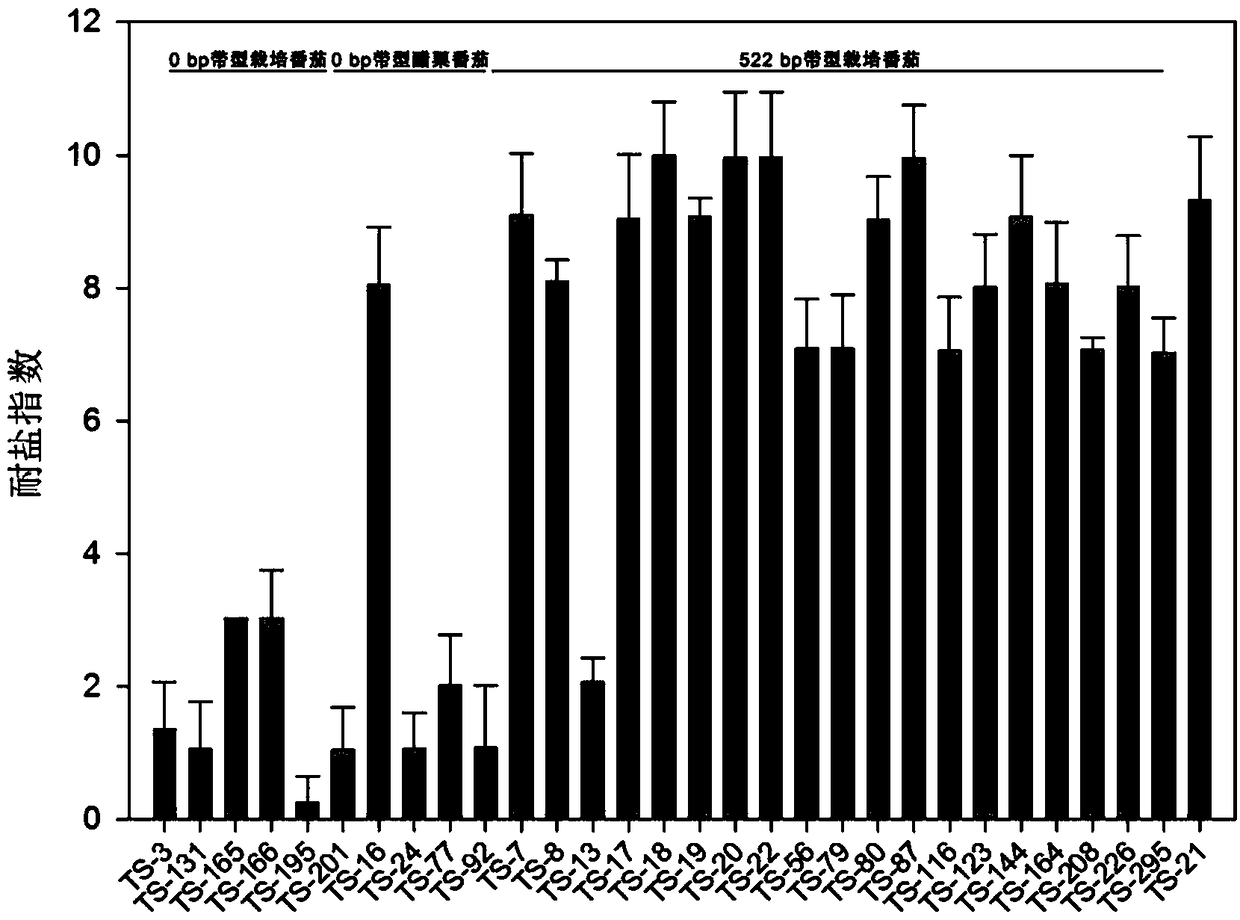
[0053] 1) Using the method in Example 2, 204 tomato germplasms (including cultivated tomato and wild tomato) were identified by PCR, and 19 parts of materials (TS-7, TS-8, TS-13, TS-17, TS-18, TS-19, TS-20, TS-22, TS-56, TS-79, TS-80, TS-87, TS-116, TS-123, TS- 144, TS-164, TS-208, TS-226, TS-295).
[0054] 2) To identify the salt tolerance of the materials obtained above, at the same time, 5 copies of cultivated tomato (TS-131, TS-165, TS-166, TS-195, TS-201) and 4 copies of Gooseberry tomato (TS-16, TS-24, TS-77, TS-92) was used as a control. The specific steps are:
[0055] (1) The plants of each salt-tolerant material screened by molecular markers were planted in sprout trays, with 8 plants for each material, randomly distributed, and 3 replicates. The plants were grown in a greenhouse with a temperature of 25±2°C, a photoperiod of 16h light / 8h dark, a light intensity o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com