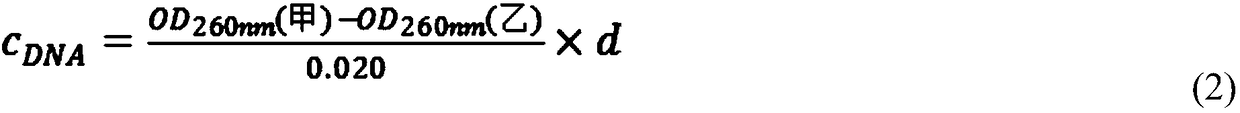Fidelity storage method for yeast DNA (Deoxyribonucleic Acid)
A yeast and fidelity technology, applied in the field of DNA material storage research, can solve the problems of low fidelity, unrealistic large-scale information storage, inconvenient access, etc., achieve high fidelity, solve large-scale information storage Effects with little problem, impact or damage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
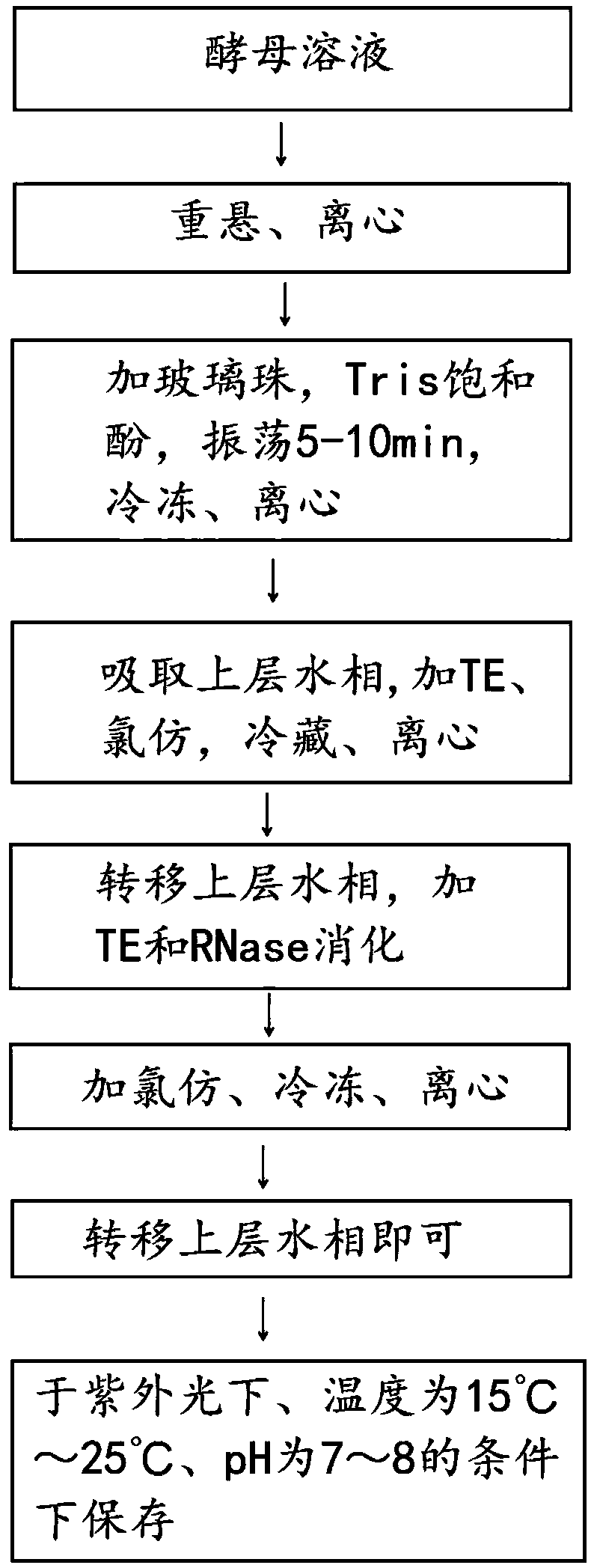
[0014] 1. Bead milling method to extract the DNA in the sample: take 4ml of active yeast liquid in a 5ml centrifuge tube, centrifuge at 8000g for 6min, resuspend the bacteria with 1600ul STE buffer, and wash the bacteria by centrifugation twice under the same conditions; the collected bacteria After suspending with 800ul TE buffer, add about 100mg of glass beads with a diameter of 425-600um, and then add 400ul Tirs saturated phenol; the above mixture is shaken on a vortex shaker for 300s, then placed in a refrigerator at 4°C for 6min, centrifuged at 13000g 6min; transfer the upper aqueous phase (about 640ul) to a new centrifuge tube, add TE buffer to make up to 800ul, then add 400ul chloroform, mix well, refrigerate at 4°C for 5min, centrifuge at 13000g for 10min. Repeat this step two to three times; transfer the upper aqueous phase (about 640ul) to a new centrifuge tube, add 200ul TE and 10ul RNase (10mg / ml), digest at 37°C for 13min; add 400ul chloroform to mix, and incubate ...
experiment example
[0017] 1. Method:
[0018] 1. Quantitative measurement of extracted DNA by ultraviolet absorption method: take 2 centrifuge tubes, add the DNA nucleic acid solution extracted in step a) respectively, then add 2mL distilled water (as a blank control) to tube A, and add 2mL molybdic acid to tube B Ammonium-perchloric acid precipitant mixed evenly;
[0019] 2. Place in an ice bath (or refrigerator) for 30 minutes, centrifuge at 3000r / min for 10 minutes; dilute the supernatants of tubes A and B respectively until the optical density is between 0.1 and 1.0;
[0020] 3. Select a quartz cuvette with an optical path of 1 cm, and at a wavelength of 260 nm, its optical density OD260nm can be calculated according to the Lambert-Beer theorem (Formula 1), and the DNA content can be calculated according to Formula 2.
[0021]
[0022]
[0023] Among them, l is the thickness of the light passing through the object to be measured, and d is the dilution factor.
[0024] 2. Exploration ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| optical density | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com