Method for the fermentative production of l-amino acids
一种氨基酸、发酵液的技术,应用在细菌领域,能够解决没有公开核苷酸序列具体应用等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
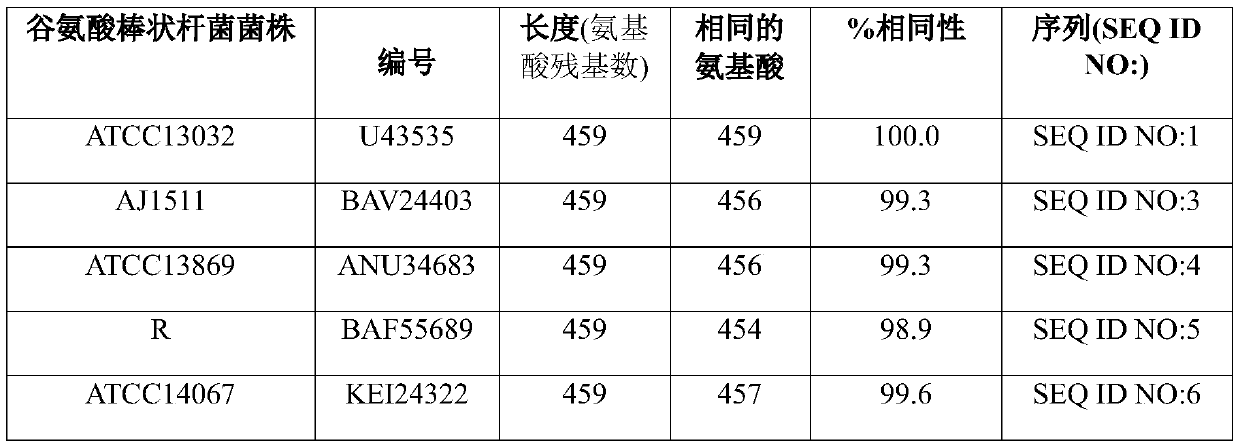
[0213] cmr gene sequences of C. glutamicum strains DM1933 and DM1797
[0214] Strain DM1933 is an L-lysine producer as described by Blombach et al. (Applied and Environmental Microbiology 75(2), 419-427, 2009). It was deposited with the DSMZ under the Budapest Treaty under the accession number DSM25442.
[0215] The nucleotide sequence of the chromosome of strain DM1933 was determined by Illumina whole genome sequencing technology (Illumina Inc., San Diego, CA, US). See, for example, Whole-Genome Sequencing for Comparative Genomics and Derivatives by Benjak et al. (2015) in (Parish T., Roberts D. (eds) Mycobacteria Protocols, Methods in Molecular Biology, Vol. 1285, Humana Press, NY, US). Article by Novo Genome Assembly and Bennet, S (Pharmacogenomics 5(4), 433-438, 2004).
[0216] The nucleotide sequence of the cmr coding sequence, including its upstream and downstream nucleotide sequences, was found to be identical to the nucleotide sequence of ATCC13032 shown in SEQ...
Embodiment 2
[0222] Construction of plasmid pK18mobsacB_Dcmr
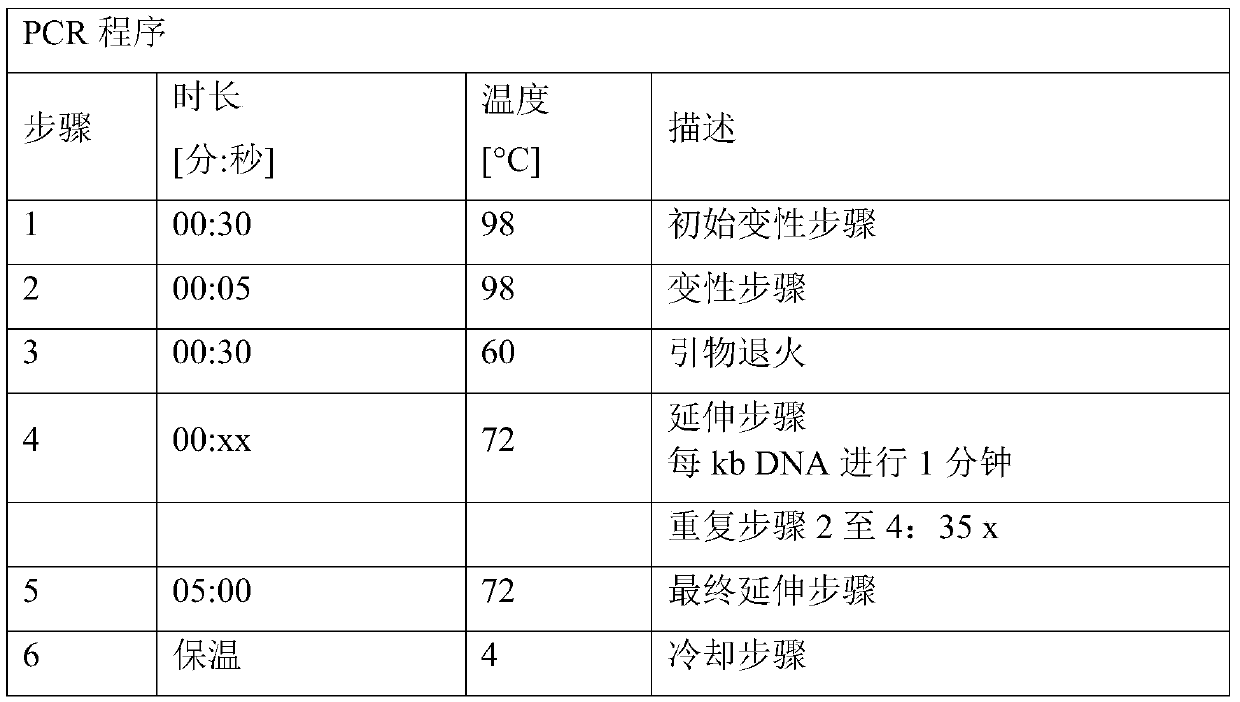
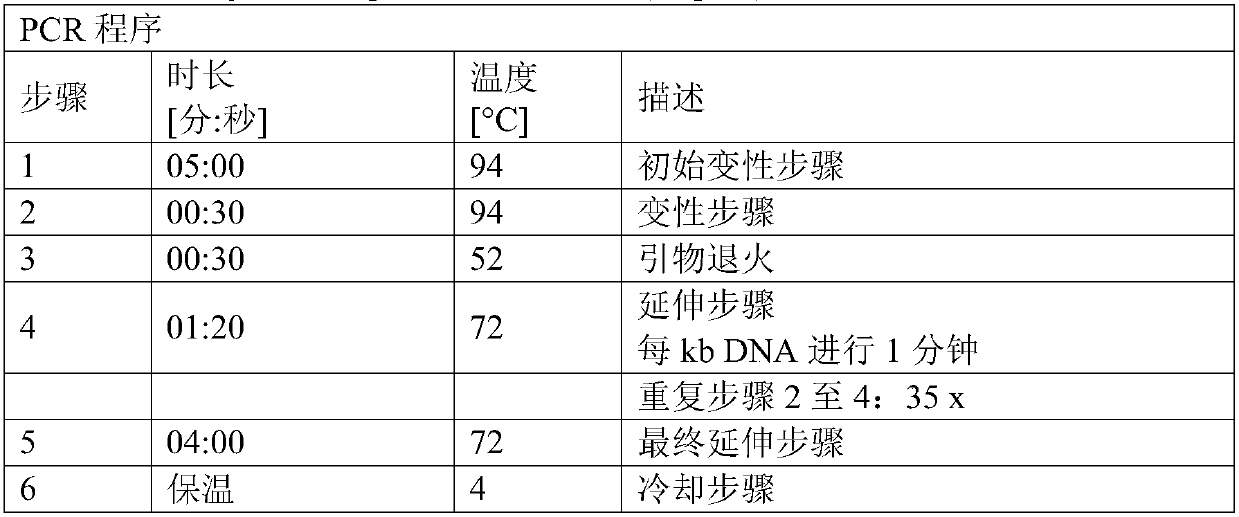
[0223] Plasmid pK18mobsacB_Dcmr was constructed to allow the introduction into the chromosome of the desired C. glutamicum strain of a deletion comprising the cmr coding sequence and its adjacent stop codon and insertion of a restriction enzyme EcoRV recognition site. The plasmid is based on The mobile vector pK18mobsacB described by et al. (Gene 145, 69-73, 1994)). pK18mobsacB_Dcmr was constructed using the Gibson Assembly method.
[0224] For this purpose, three polynucleotides or DNA molecules were generated separately: one called cmr_up containing the upstream sequence (5'-flanking sequence) and a second called cmr_down containing the downstream sequence of the cmr coding sequence (3'-flanking sequence) polynucleotide. The third polynucleotide was plasmid pK18mobsacB linearized with the restriction endonuclease Xbal. Polynucleotides cmr_up and cmr_down were fused during the Gibson Assembly process to generate polynucle...
Embodiment 3
[0242] Construction of strain DM1933_Δcmr::EcoRV
[0243] The plasmid pK18mobsacB_Dcmr was used to introduce the deletion of the entire cmr coding sequence and its adjacent stop codon into the chromosome of L-lysine producing strain DM1933, accompanied by the insertion of the recognition site for the restriction endonuclease EcoRV.
[0244] The deletion of the entire cmr coding sequence with its adjacent stop codon and the concomitant insertion of a recognition site for the restriction enzyme EcoRV is abbreviated as Δcmr::EcoRV or deltacmr::EcoRV.
[0245] Chemically competent cells of E. coli strain S17-1 were transformed with the plasmid DNA of pK18mobsacB_Dcmr obtained in Example 2. Materials and methods described in The modified conjugation method of et al. (Journal of Bacteriology 172, 1663-1666, 1990) was used for conjugative transfer into strain DM1933 and for selection of transzygote clones by their sucrose resistance and kanamycin sensitivity phenotypes.
[0246]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com