DNA-binding protein using PPR motif and use of said DNA-binding protein
A protein and motif technology, applied in the protein field
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0448] [Example 1: Collection of novel dPPR molecules]
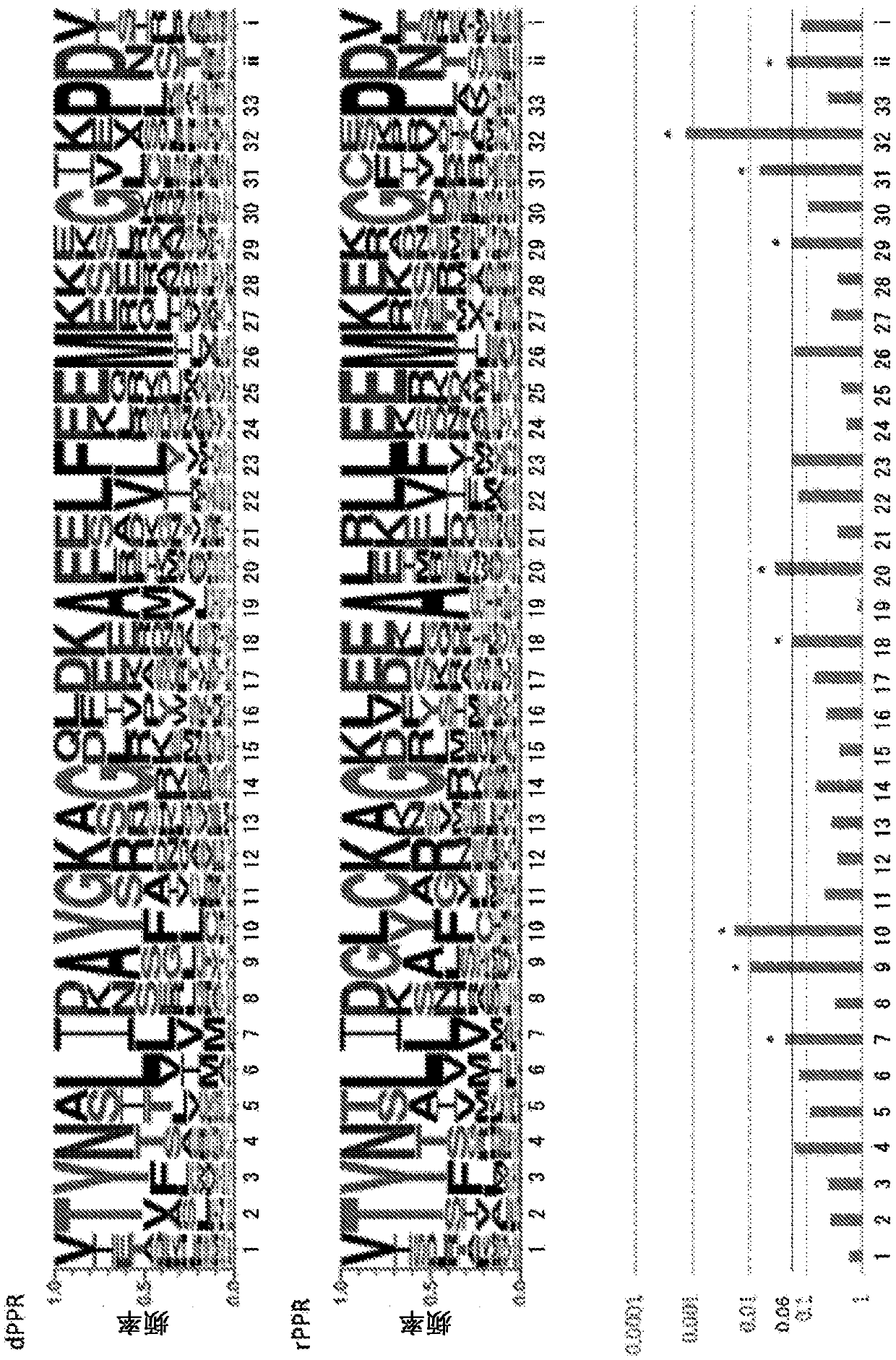
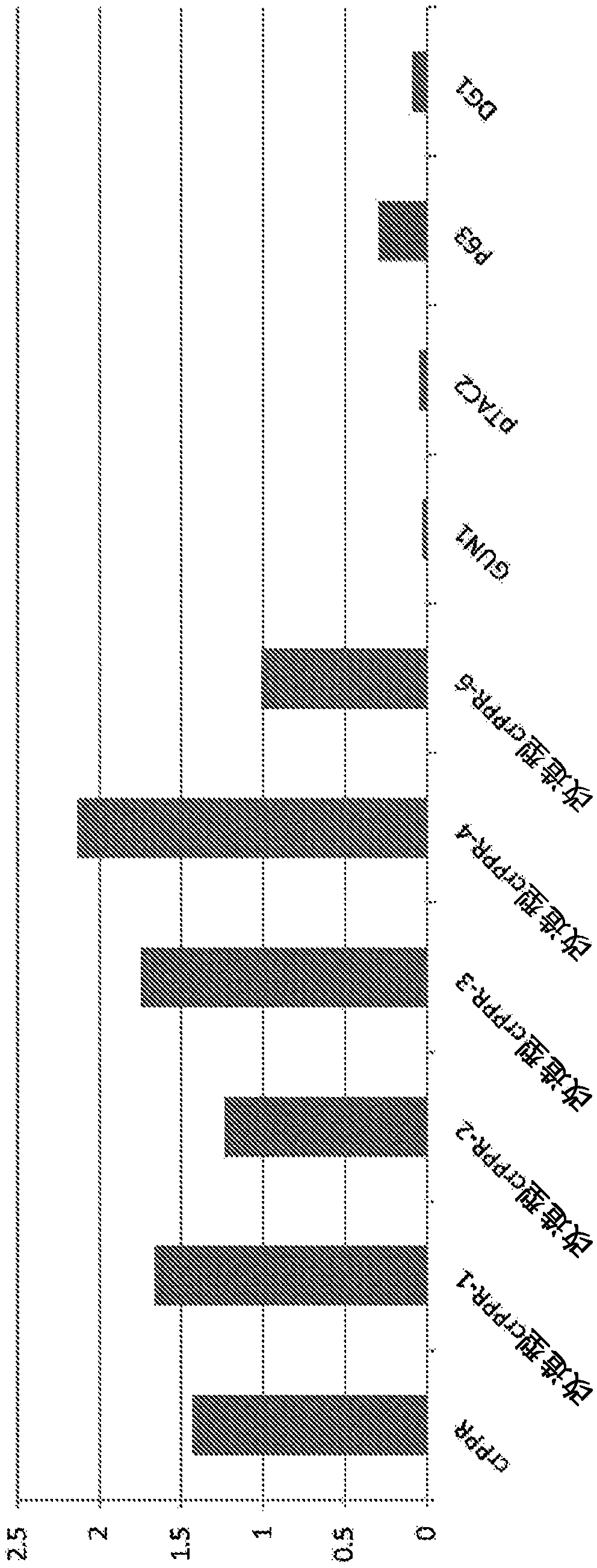
[0449] Known dPPR proteins are only P63, GUN1, pTAC2, GRP23, and DG1 described in the previous patent (the above-mentioned Patent Document 4), and it is difficult to obtain information for the generalization and improvement of artificial nucleic acid binding modules based on PPR technology. Therefore, this time, we decided to screen for PPR proteins with DNA-binding ability to expand dPPR proteins. The genes of the dPPR molecules found by chance so far contain introns, but the rPPR genes hardly contain introns at all. Using this as an indicator, the whole genome sequence of Arabidopsis thaliana, which is a model plant, was analyzed. As a result, there were 42 PPR genes containing two or more introns. In this example, the DNA-binding ability of these 42 potential dPPR molecules was analyzed in an attempt to identify new dPPR molecules.
[0450] (experimental method)
[0451] 1. Construction of dPPR expression vector
...
Embodiment 2
[0474] [Example 2: dPPR motif-specific amino acid sequence analysis]
[0475] Based on the amino acid sequence information of the modules contained in the dPPR protein identified in Example 1, the dPPR motif-specific amino acid sequence was analyzed.
[0476] First, 9 dPPR proteins were selected from the 18 dPPR proteins identified in Example 1 in order to match to some extent the motif numbers of rPPR proteins in the F-test. Specifically, for dPPR proteins, the DNA binding force was compared with OTP80 by T-test, and dPPR proteins were classified into 3 groups of 0.05-0.01, 0.01-0.001, and <0.001 based on the obtained values, and randomly selected from each group 3, so choose 9 kinds. At all positions, compare the occurrence frequency of amino acids in the PPR motifs (recorded in the order of 1, 2, 3...) of the 9 dPPR molecules and the 5 known rPPR molecules in the table below, and try to determine the characteristics of the dPPR protein The location of the active amino aci...
Embodiment 3-1
[0487] [Example 3-1: Establishment of artificial nucleic acid binding module construction method based on dPPR motif-specific amino acid sequence 1]
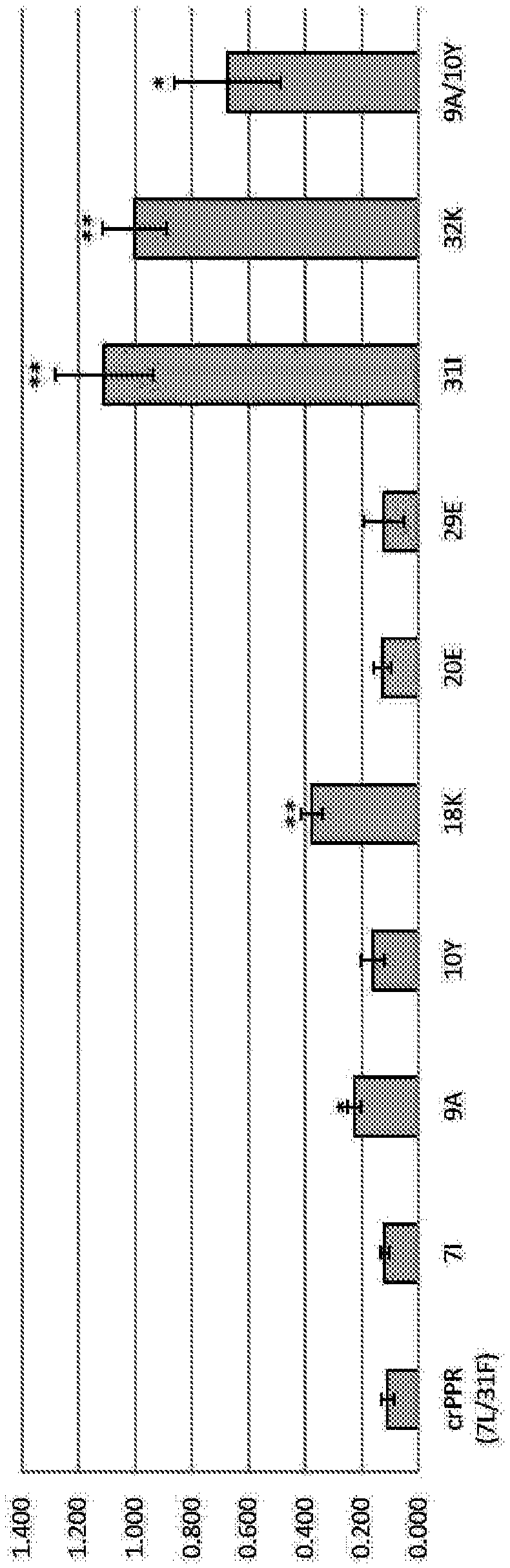
[0488] In this Example, in order to verify whether the DNA-binding ability of the PPR protein is enhanced by the dPPR-specific amino acid sequence, the DNA-binding ability of the modified rPPR incorporating the dPPR-specific amino acid sequence was investigated. As the basic rPPR, the consensus PPR (cPPR) reported in Non-Patent Document 15 (Coquille et al., 2014, An artificial PPR scaffold for programmable RNA recognition) was used. It should be noted that cPPR is known as an RNA-binding protein (hence it may be expressed as crPPR), but it is not known whether it binds to DNA. The modification of crPPR utilizes Genewiz's gene synthesis. The DNA binding ability of the engineered crPPR was analyzed by the method used in Example 1. It should be noted that the target sequence of crPPR is AAAAAAAA.
[0489] It should be noted that...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com