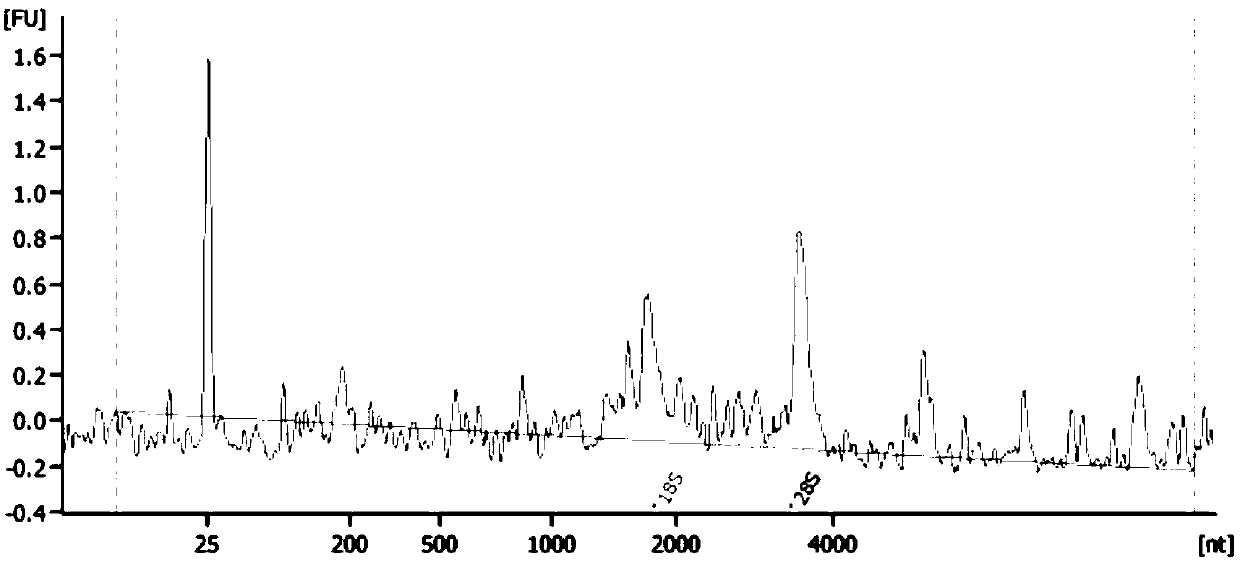
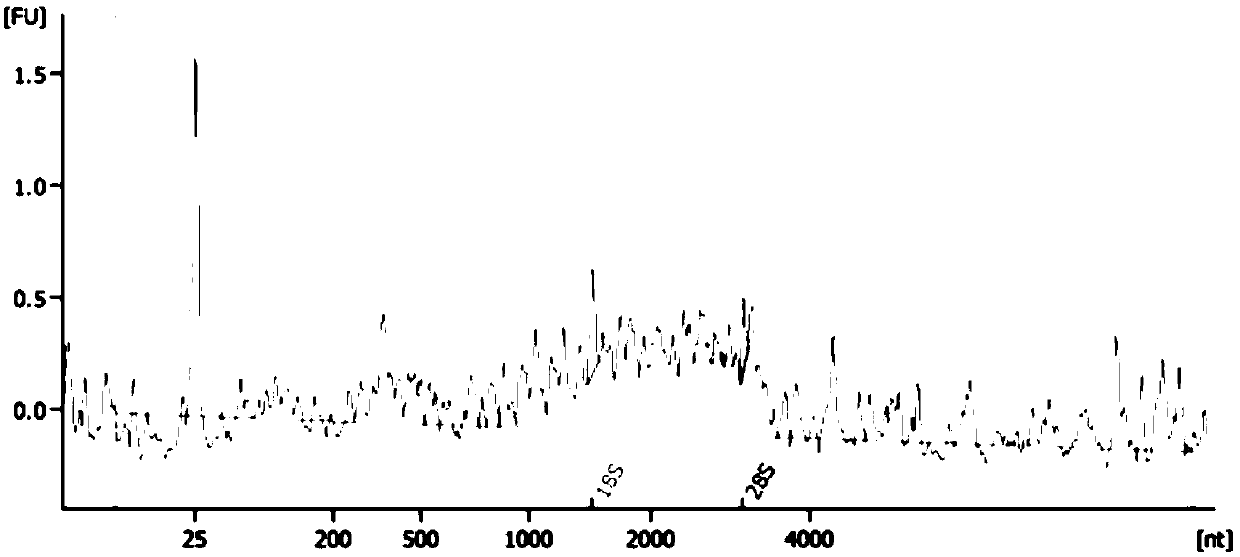
Method for separating cell nucleus from human frozen tumor tissue suitable for single cell sequencing
A tumor tissue and cell nucleus technology, applied in the field of sequencing, can solve the problems of lack of quality control results display instructions, etc., achieve the effect of simple reagent consumables and increase recovery rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] (1) Reagent preparation
[0063] 1. NST lysis buffer (prepare the following two components separately and combine and mix well):
[0064] 1) 80mL of NST: 146mM NaCl, 10mM Tris base at pH 7.8, 1mM CaCl 2 ,21mMMgCl 2 ,0.05%BSA,0.2%Nonidet P-40);
[0065] 2) 20mL of 106mM MgCl 2 , 5mM EDTA, 1mg DAPI;
[0066] 2. Nuclei Wash and Resuspension Buffer (Nuclei W&R Buffer):
[0067] 1×PBS, 1.0% BSA, 0.2U / μL RNase inhibitor.
[0068] (2) Tissue cell lysis
[0069] 1. Pre-cool the prepared NST / DAPI lysis buffer and Nuclei W&R Buffer in a refrigerator at 4°C;
[0070] 2. Put the plastic petri dish on ice to pre-cool, add 3mL NST / DAPI lysis buffer;
[0071] 3. Put ~30mg of tissue into a Petri dish, cut it into pieces smaller than 1mm3 with a scalpel blade, place it on ice for 15 minutes, and shake gently every 5 minutes to mix;
[0072] 4. Disperse the tissue by blowing and aspirating 10-15 times, and filter it into a 15mL tube with a 37μm cell strainer;
[0073] (3) Nucle...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com