Boehmeria nivea leaf back velutinous main effect QTL and closely linked molecular marker and application thereof
A ramie leaf and marking technology, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problems of reduced feeding performance and poor palatability of ramie, and achieve the effect of good technical support
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] The acquisition of the SNP marker of ramie leaf dorsal felt wool is as follows:
[0069] 110 single-plant RaF2 populations were obtained by crossing the parent Zhongsizhu 1 with Qingye Ramie, and the 110 RaF2 populations and the transcriptomes of the two parents were sequenced;
[0070] A high-density genetic map containing 1085 molecular markers was constructed by Jionmap4.0 software, and the 1085 markers were evenly distributed on 14 linkage groups, with an average marker spacing of 1.95cM;
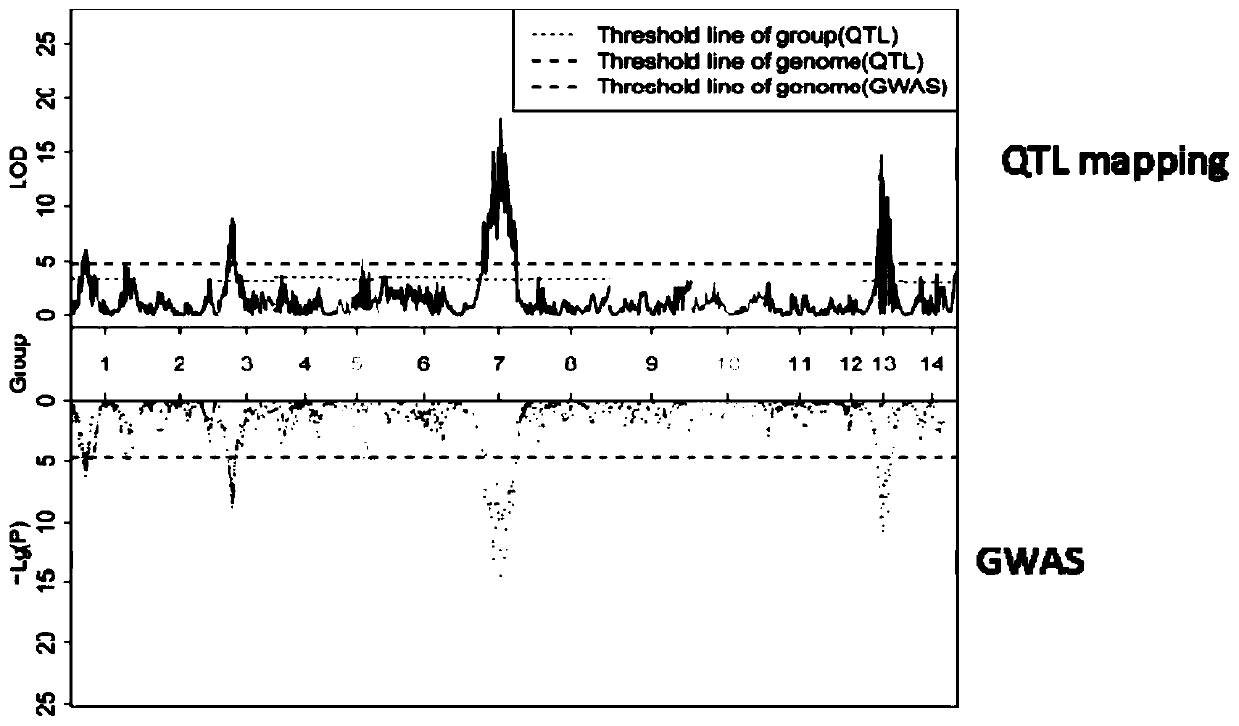
[0071] According to the RaF2 phenotype data, combined with the high-density linkage map, the traits were detected by MapQTL (version: 6.0) software, and the 1085 SNP sites on the integrated genetic map were compared with 2 by using plink (version: 1.07) software through GWAS. The association analysis of a single SNP locus is carried out for each trait, and the results are as follows figure 1 shown.
[0072] figure 1 , the upper half of the figure shows the LOD values obtaine...
Embodiment 2
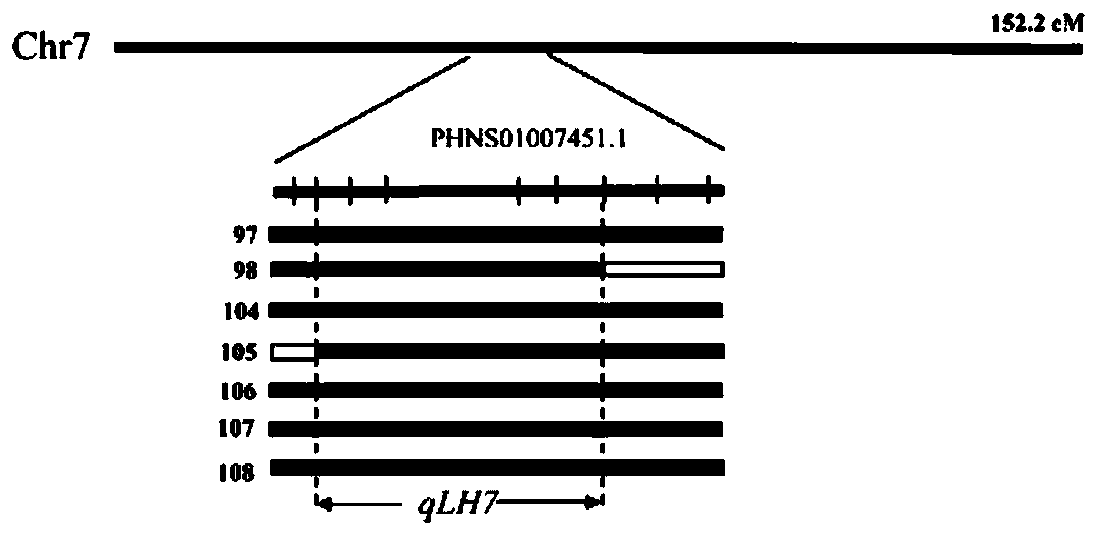
[0099] Based on the SNP marker of ramie leaf dorsal hair on chromosome 7 of ramie, the SSR and INDEL markers of ramie leaf dorsal hair were developed based on the scaffold sequence of chromosome 7, and then the SSR and INDEL primers were designed with PRIMER5 and DNAMAN. 16 pairs of SSR primers and 7 pairs of INDEL primers were synthesized by a biological company.
[0100] The genotype identification of 110 individual RaF2 populations obtained from crossing Sizhu 1 and Qingye Ramie in the parent, the process includes the following steps:
[0101] 1) DNA extraction work;
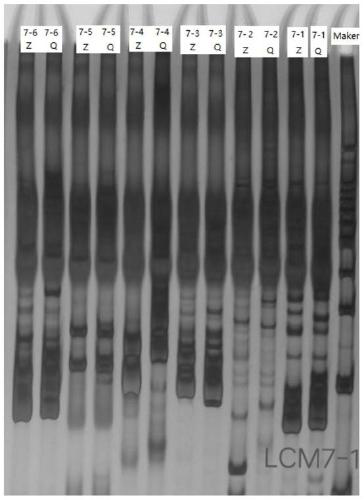
[0102] 2) Use 16 pairs of synthetic SSR primers and 7 pairs of INDEL primers to perform PCR amplification on the genome of each individual plant, and then analyze the bands by electrophoresis for genotype analysis. The genotype consistent with Zhongsizhu No. 1 is marked as "Z", and the same genotype as Qingye Ramie is marked as "Q". If a single plant contains both Zhongsizhu 1 and Qingye Ramie genotypes, it ...
Embodiment 3
[0115] Based on the SNP marker of ramie leaf dorsal hair on chromosome 13 of ramie, SSR and INDEL markers were developed based on the scaffold sequence of chromosome 13, and then primers for SSR and INDEL were designed with PRIMER5 and DNAMAN. 17 pairs of SSR primers were synthesized by a biological company.
[0116] The genotype identification of 110 individual RaF2 populations obtained from crossing Sizhu 1 and Qingye Ramie in the parent, the process includes the following steps:
[0117] 1) DNA extraction work;
[0118] 2) Using SSR primer 17 to perform PCR amplification on the genome of each individual plant, and then analyze the bands by electrophoresis for genotype analysis. The same genotype of Zhongsizhu No. 1 is marked as "Z", and the same genotype of Qingye Ramie is marked as "Q". If a single plant contains both Zhongsizhu 1 and Qingye Ramie genotypes, then The heterozygous genotype is marked as "H".
[0119] Some electrophoresis results such as Figure 5-7 shown...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com