Method for detecting EGFR L858R locus in lung cancer by ddPCR and application thereof
A technology of L858R and detection method, which is applied in ddPCR detection and application field of EGFR L858R locus in lung cancer, which can solve the problems of complex procedures, complex positive interpretation methods, low sensitivity, etc., and achieve the effect of high detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0086] Embodiment 1: The method for detecting EGFR L858R mutation by ddPCR method;
[0087] 1) Screening of primer pairs;
[0088] According to the sequence of exon 21 of EGFR, design 5 pairs of primers (primers were synthesized from Shanghai Sangon Biotechnology Co., Ltd.), the specific sequences are as follows:
[0089]
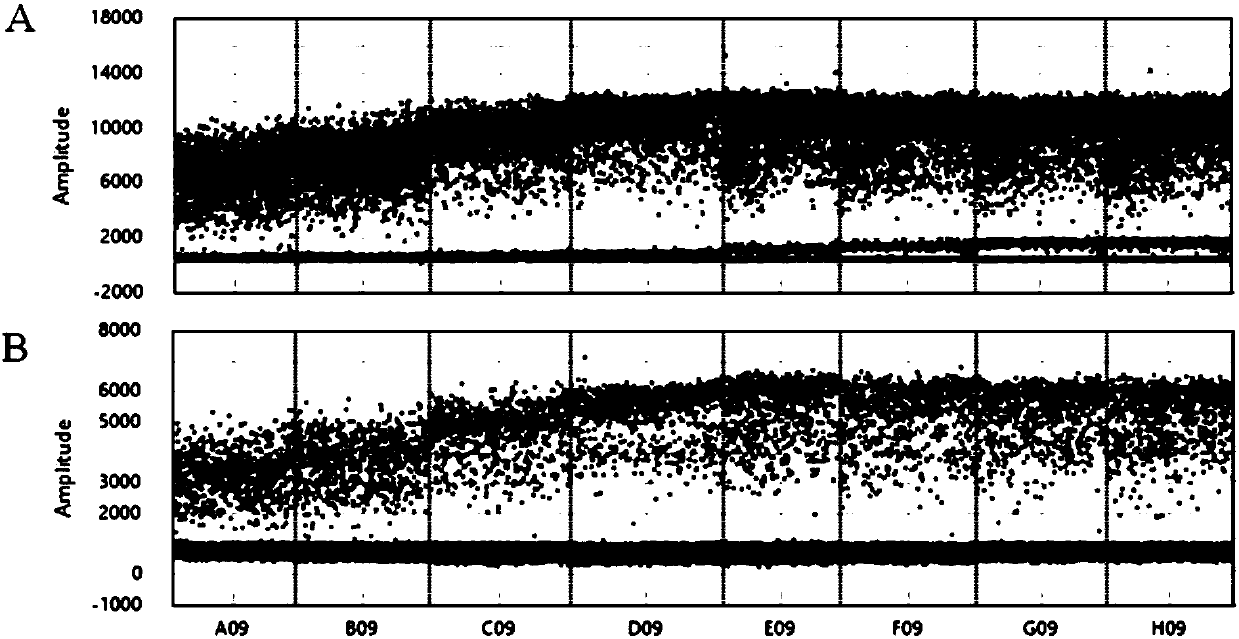
[0090] Screening PCR system: 0.2ul of Taq HS polymerase, 2ul of 10×HS PCR buffer, 1.6ul of dNTP mixture, 1ul of primer 1, 1ul of primer 2, 5ul of tgDNA (DNA template), and make up the system to 20ul with water. Screening PCR program: 95°C for 10 min, 30 cycles (94°C for 30s, 55°C for 30s, 72°C for 20s). After PCR screening, primer pair 1 (1-858-F / 1-858-R) had the highest amplification efficiency for the target fragment and did not produce primer dimers. The primer pair is used as a detection primer pair for the L858R mutation of the EGFR gene.
[0091] 2) Optimization of the ddPCR reaction program;
[0092] According to the EGFR gene L858R mutation s...
Embodiment 2
[0101] Embodiment 2; ddPCR method to detect the method verification of EGFR L858R mutation;
[0102] According to the result of ddPCR using 1975 as a template in Example 1, the copy number of the FAM channel and the total copy number of 1975 were calculated, the two were divided and multiplied by 100%, and the mutation rate of 1975 or the ratio of mutant template was 66%.
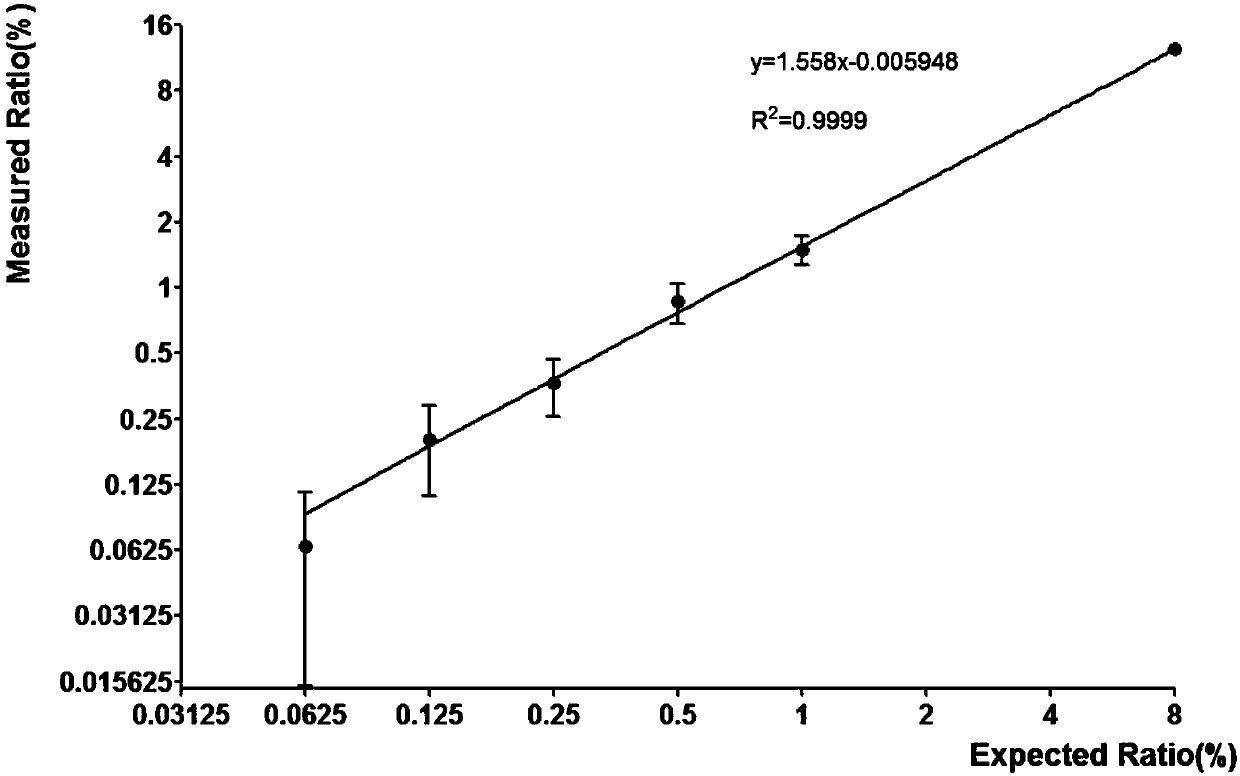
[0103] The genome copy numbers of 1975 and tgDNA were calculated according to Example 1, and both were diluted to 4000 copies / ul with TE. 1975 was incorporated into tgDNA so that the ratios of the L858R mutation templates were 0.0625%, 0.125%, 0.25%, 0.5%, and 1%, respectively, and the gradient dilution templates were made.
[0104] Validate the L858R mutation detection method as follows. Verify the ddPCR system: 2×ddPCR master mix (without dUTP) 11ul, primer 1 (1-858-F) 1.1ul, primer 2 (1-858-R) 1.1ul, probe 1 (WTp-L858R) 0.55ul , probe 2 (MTp-L858R) 0.55ul, template 5.5ul, add water to make up to 22ul. ...
Embodiment 3
[0107] Example 3: The L858R mutation detected by the ddPCR method guides the administration of tumor-targeted drugs;
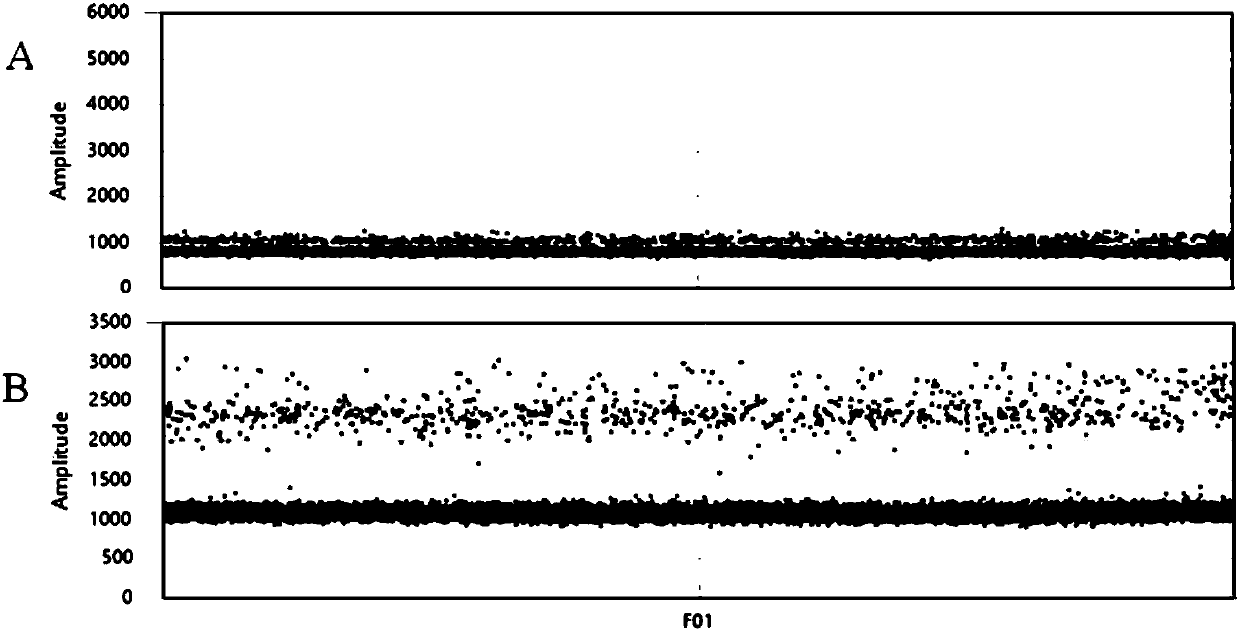
[0108] Cases TP009 and TP034 were stage IV patients with lung adenocarcinoma. By collecting venous blood, centrifuge twice to separate plasma (the first low-speed centrifugation, 4°C, 1900g, 10min, the second high-speed centrifugation, 4°C, 16000g, 10min), and carefully pipette 1ml of the supernatant into a 50ml centrifuge tube. Plasma free nucleic acid was extracted by QIAGEN free nucleic acid extraction kit. The extracted nucleic acid was quantified by Qubit. After quantification, it was stored in a -20°C refrigerator in the sample preparation room.
[0109]The ddPCR method of the present invention detects the EGFR L858R mutation. Detection PCR system: 2×ddPCR master mix (without dUTP) 11ul, primer 1 (1-858-F) 1.1ul, primer 2 (1-858-R) 1.1ul, probe 1 (WTp-L858R) 0.55ul , probe 2 (MTp-L858R) 0.55ul, template 5.5ul, add water to make up to 22ul.
[0110] ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com