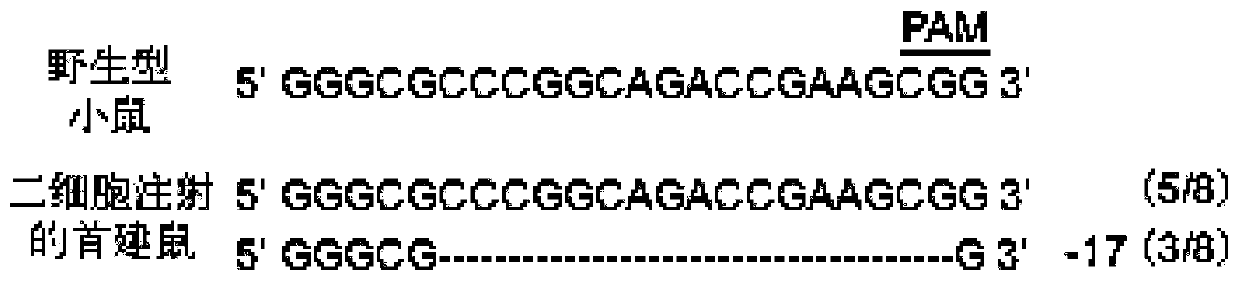Method for constructing knock-out mouse model to analyze lethal gene function by utilizing CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR associated protein 9) system
A technology of model analysis and gene function, applied in the biological field, can solve the problems of inability to quickly realize lethal genes, cumbersome production steps, unfavorable research development, etc., and achieve the effect of long production cycle, cumbersome production steps and low operation difficulty
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
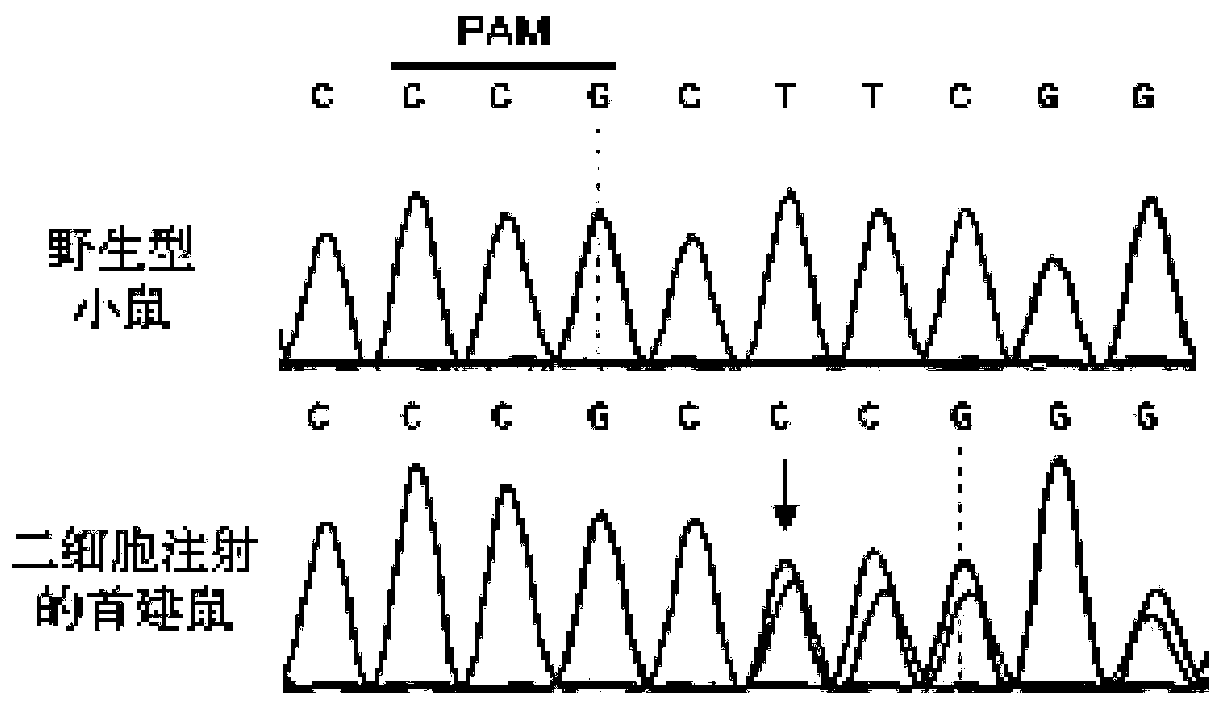
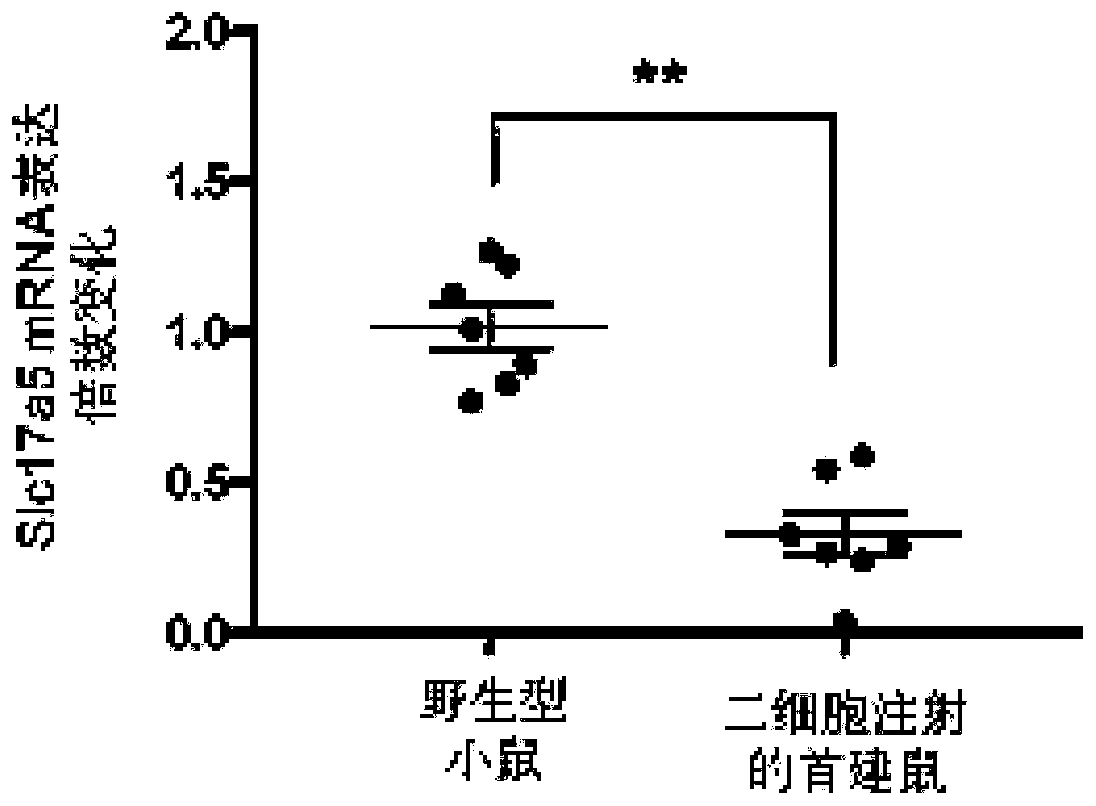
[0056] A method of using the CRISPR / Cas9 system to construct a systemic knockout mouse model of the postnatal lethal gene Slc17a5 that can be subcultured by microinjection of two-cell embryos. The present invention will be further described below in conjunction with specific examples.
[0057] 1.1 Construction of CRISPR-Cas9 system components
[0058] 1.1.1 Design of sgRNA sequence for gene knockout
[0059] (1) Enter the NCBI website and query the Gene ID of mouse Slc17a5: 235504.
[0060] (2) Enter https: / / portals.broadinstitute.org / gpp / public / analysis-tools / sgrna-design website, enter Gene ID to find available sgRNA.
[0061] (3) Select the sgRNA according to the score given by the website. The higher the score, the lower the off-target effect and the better the specificity of the sgRNA. At the same time, the position of the sgRNA on the CDS should also be considered comprehensively. Use the domain architecture retrieval tool of NCBI to predict which important structural ...
Embodiment 2
[0196] A method of using the CRISPR / Cas9 system to construct a subcultured embryonic lethal gene Virma systemic knockout mouse model by microinjection of two-cell embryos. The present invention will be further described below in conjunction with specific examples.
[0197] 2.1 Construction of CRISPR-Cas9 system components
[0198] For detailed steps, refer to the method in 1.1, Gene ID of Virma: 66185. sgRNA sequence, sgRNA1 sequence is 5'-CUAUGGGCUCGUACUCCCGG-3' (SEQ ID NO.2).
[0199] Oligodeoxynucleotide (DNA oligo, 5'→3') synthesized by Sangon Biotech
[0200] sgRNA1F: taatacgactcactatagCTATGGGCTCGTACTCCCGGgtttaagagctatgctggaaa (SEQ ID NO. 7);
[0201] sgRNAR:aaaagcaccgactcggtgcc (SEQ ID NO.4)
[0202] Finally, the in vitro transcription and purification of Virma sgRNA1 and Cas9 mRNA were completed.
[0203] 2.2 Two-cell embryo microinjection and embryo transfer in mice
[0204] Detailed steps are with reference to the method of step 1.2 in embodiment 1
[0205] Virm...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com