High-throughput functional gene screening method and system for quantitative analysis of cell phenotype images
A functional gene and quantitative analysis technology, applied in image analysis, material analysis, sequence analysis, etc., can solve the problems of low throughput and inability to accurately quantify cell phenotypes, achieve good repeatability, fast recognition speed, and reduce workload Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
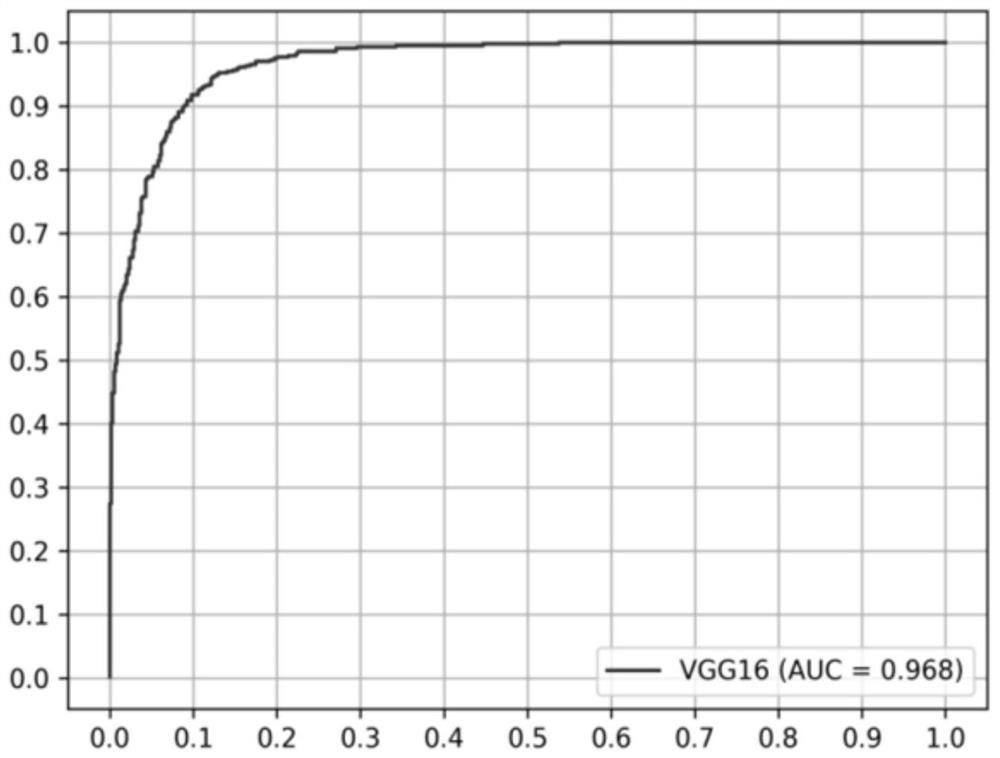
Image
Examples
Embodiment 1
[0042] The 103 phosphorylation regulator genes in the iEKPD database were screened to see whether they were related to autophagy.
[0043] 1. Generate a training set: import GFP-Atg8 into yeast cells to express and produce green fluorescence, use FM4-64 to mark vacuoles to produce red fluorescence, and the conditions for phenotype generation include nitrogen reduction and rapamycin induction (rapamycin induction) Wait. The cells were non-autophagy phenotype cells at 0 hours, and the cells underwent autophagy after 2 hours under reduced nitrogen conditions. Generate large numbers of images of phenotyped and non-phenotyped cells using fully automated fluorescence microscopy.
[0044] 2. Image processing: enhance the fluorescence signal by enhancing the contrast and deconvolution and denoising. Deconvolution and denoising: use PSF (point spread function) of different sizes to restore blurred images, analyze and reconstruct PSF, so as to improve the restored image.
[0045] 3. ...
Embodiment 2
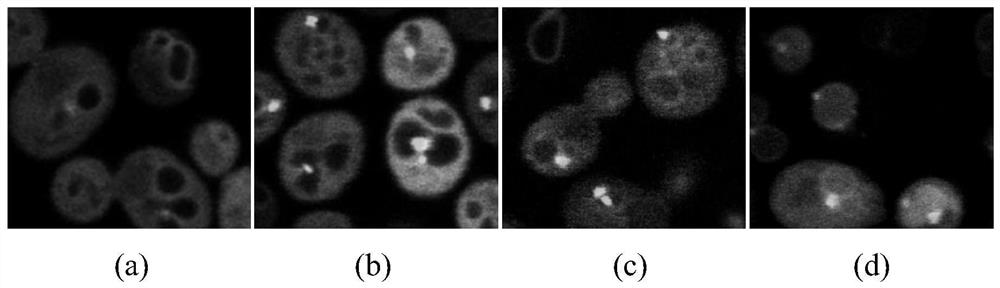
[0060] figure 1 (a) Cell phenotype at 0h after atg1 knockout under nitrogen reduction condition, no autophagy at this time; figure 1 (b) Cell phenotype at 2 hours after atg1 knockout under nitrogen reduction condition, at which point autophagy did not occur; figure 1 (c) is the cell phenotype at 0h after SNF1 is knocked out under nitrogen reduction condition, and there is no autophagy at this time; figure 1(d) Cell phenotype at 2 h after SNF1 knockout under nitrogen reduction condition, at which time autophagy occurs. like figure 1 As shown in (d), GFP-Atg8 enters FM4-64-labeled vacuoles during autophagy; as figure 1 (a), figure 1 (b) and figure 1 As shown in (c), GFP-Atg8 is mainly localized outside the vacuole when cells are not autophagy. We can teach computers to recognize this phenotypic difference to distinguish autophagic from non-autophagic cells.
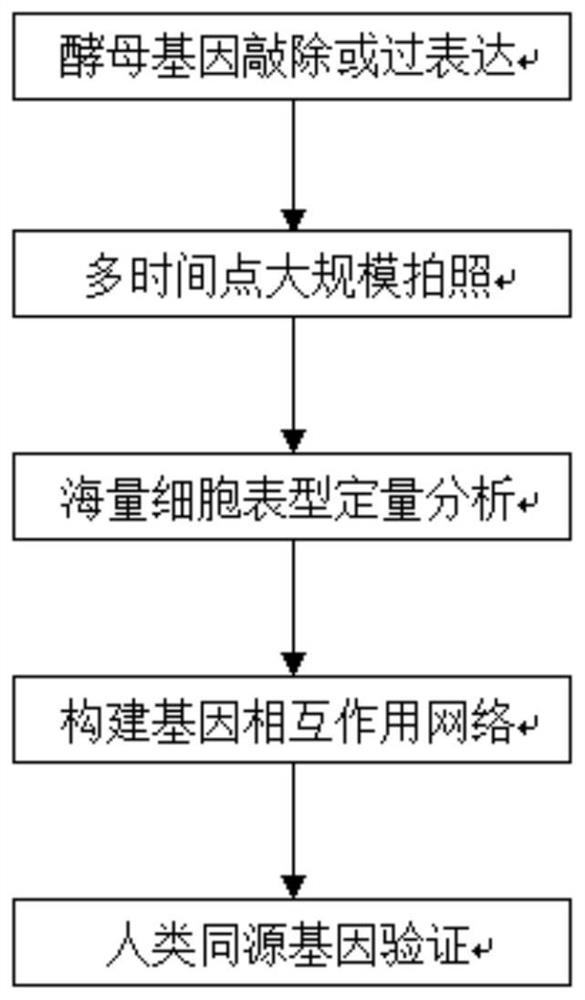
[0061] figure 2 It is a flowchart of a high-throughput gene screening method based on quantitative analysis of m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com