RPA primer, probe and kit for detecting shell parasitic perkinsus
A technology of sending Qin worms and a kit, which is applied in the field of rapid diagnosis of aquatic animal parasites, can solve problems such as ineffective drug treatment and inability to guarantee drug concentration, and achieve the effects of visualization of test results, great potential for promotion and application, and simple operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Embodiment 1 Primer and probe design
[0038] Primer design: Using the ribosomal DNA internal transcriptional spacer sequences of the seven Piedia species discovered so far as design templates, a pair of primers that can amplify all Piedias were designed and screened. The primer sequences are:
[0039] Upstream primer F1: 5'-CGATGAAGGACGCAACGAAGTG-3' (SEQ ID NO.1);
[0040] Downstream primer R1: 5'-CAAGCGGGATACAAAGCATTAGATT-3' (SEQ ID NO.2).
[0041] A probe is designed in the amplified target sequence, and the probe Probe sequence is: 5'-CAGAATTCCGTGAACCAGTAGAAAATCTCAACGCATACTGCACAAAGGGGA-3' (SEQ ID NO.3).
[0042] In order to directly detect the results with test strips, a biotin label was added to the 5' end of the primer R1, a FAM label was added to the 5' end of the probe, dSpacer or THF was added in the middle, and a C3Spacer label was added to the 3' end.
[0043] The sequence after fluorescent labeling is as follows:
[0044] R1': 5'-biotin-CAAGCGGGATACAAAGCA...
Embodiment 2
[0049] The present embodiment provides the method that adopts RPA method to detect shellfish parasitic Pythias, comprising the following steps:
[0050] (1) Refer to Example 1 for primer design.
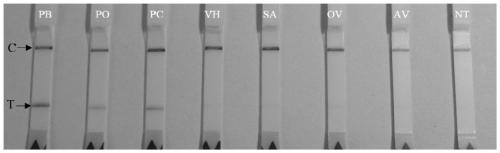
[0051] (2) DNA extraction: DNA from gill tissues of different shellfish is extracted for the detection of Piedia worms.
[0052] (3) Constant temperature amplification of recombinase polymerase: use the DNA of shellfish gill tissue as a template, use F1, R1' as upstream and downstream primers, and Probe' as probe to carry out constant temperature amplification. The amplification system is 50 μL, and the Rehydration 29.5 μL of buffer, 2.1 μL of upstream and downstream primers (10 μM), 0.6 μL of probe, 12.2 μL of sterilized ultrapure water, and 1 μL of DNA sample, a total of 47.5 μL of the system was pre-mixed, transferred to a reaction tube, and then 2.5 Add μL of MgAc (280mM) solution to the corresponding tube cap, mix well, and let it react at room temperature for 20-25min.
[0053]...
Embodiment 3
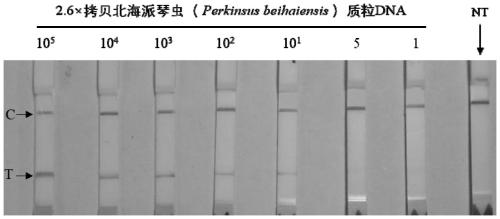
[0055] Sensitivity experiment of the diagnostic method of the present invention
[0056] (1) Amplify the ITS of Perkinsus beihaiensis using the general primers Perkinsus beihaiensis recommended by OIE Sequence, prepare the recombinant plasmid of this target sequence, and extract the plasmid DNA of the North Sea Piedia worm.
[0057] (2) Dilute the DNA concentration (copies / μL) of the North Sea Piedia worm plasmid to 2.6×10 5 , 2.6×10 4 , 2.6×10 3 , 2.6×10 2 , 2.6×10 1 , 2.6×5, 2.6×1, as DNA standards for sensitivity analysis.
[0058] (3) Using different concentrations of Beihai Paiqinia plasmid DNA as a template, using F1, R1' as upstream and downstream primers, and Probe' as a probe for constant temperature amplification, the amplification system is 50 μL, first rehydration buffer 29.5 μL, upper 2.1 μL of each downstream primer (10 μM), 0.6 μL of probe, 12.2 μL of sterilized ultrapure water, and 1 μL of DNA sample, a total of 47.5 μL of the system was pre-mixed, transf...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com