Double-gene recombinant complex macrophage vaccine and preparation method and application thereof
A macrophage and complex technology, which is applied in the field of vaccines prepared by the same and its preparation, and dual-gene recombination complexes, can solve the problems of many influencing factors, unfavorable target gene expression and subsequent treatment, difficult evaluation and analysis of experimental results, and the like, Achieve the effect of inhibiting inflammatory response and brain edema, good development and application prospects, and improving neurological function scores
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] The preparation of embodiment 1 double gene recombination compound
[0044] 1.1 Cloning of IRAK2RNAi
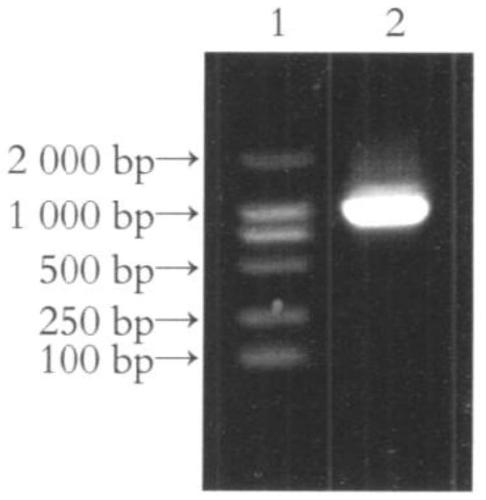
[0045] According to the instructions of the Trizol kit, the total RNA of human liver tissue was extracted, and the total cDNA was obtained by reverse transcription. Then, using the total cDNA as a template, F1:5'-aat ctgcag acttttcgcccgcacagagcgg-3' (SEQ ID No.3, the underlined part is the restriction site of Bgl1) and R1: 5'-tac gaattc aagcgcgtgcacgaggaggtcacg-3' (SEQ ID No.4, the underlined part is the restriction site of Apo1) is the upstream and downstream primers to amplify the full-length RNAi of IRAK2 by PCR. ; Then denature at 94°C for 45 seconds, anneal at 58°C for 60 seconds, and extend at 72°C for 1 minute, a total of 30 cycles; finally extend at 72°C for 5 minutes. The PCR product was identified by agarose gel electrophoresis, recovered and purified by gel extraction kit, and ligated with pT-Easy vector. The ligated product was transformed into Escheric...
Embodiment 2
[0060] Example 2 Preparation of double-gene recombinant complex transfection macrophage vaccine
[0061] 2.1. Sorting of macrophages
[0062] C57BL / 6 normal mice were taken, killed by neck dislocation, and routinely disinfected with 75% alcohol. Inject 5 mL of serum-free RPMI 1640 medium into the abdominal cavity of mice, rub the abdomen repeatedly for 5 minutes, let it stand for 3 minutes, open the abdominal cavity, extract the peritoneal fluid, centrifuge, detect the cell density, and plant it in a 6-well plate for 3 hours to remove unattached cells , add medium and cytokines: IFN-γ 10ng / mL, LPS 100ng / mL; M2 macrophages: IL-4 10ng / mL) to stimulate culture for 24h.
[0063] 2.2. Double-gene recombination complex transfected macrophages
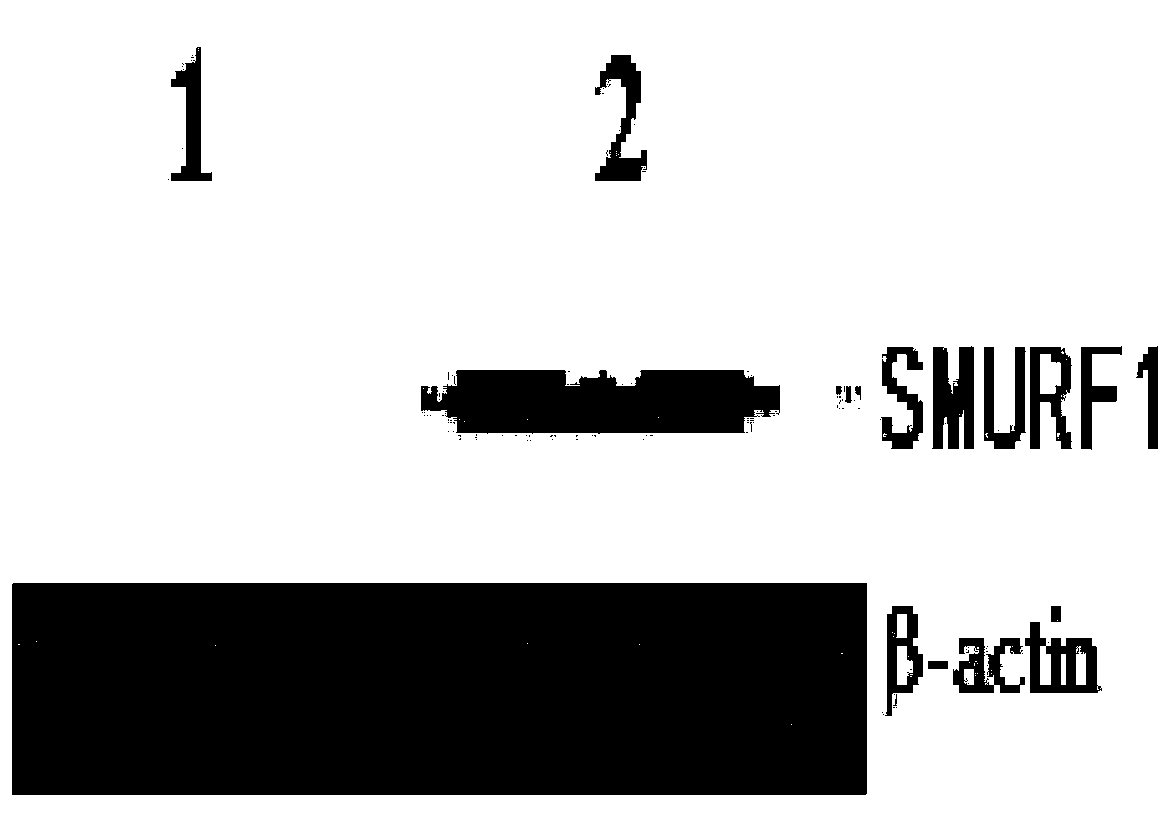
[0064] Inoculate macrophages in a 6-well plate at a cell concentration of 1×106 / ml, add the fourth-generation double-gene recombination complex at a multiplicity of infection (MOI) of 100, collect cells 48 hours after infection, wash with P...
Embodiment 3
[0068] Example 3 Detection of Cerebral Edema in Mice with Cerebral Hemorrhage
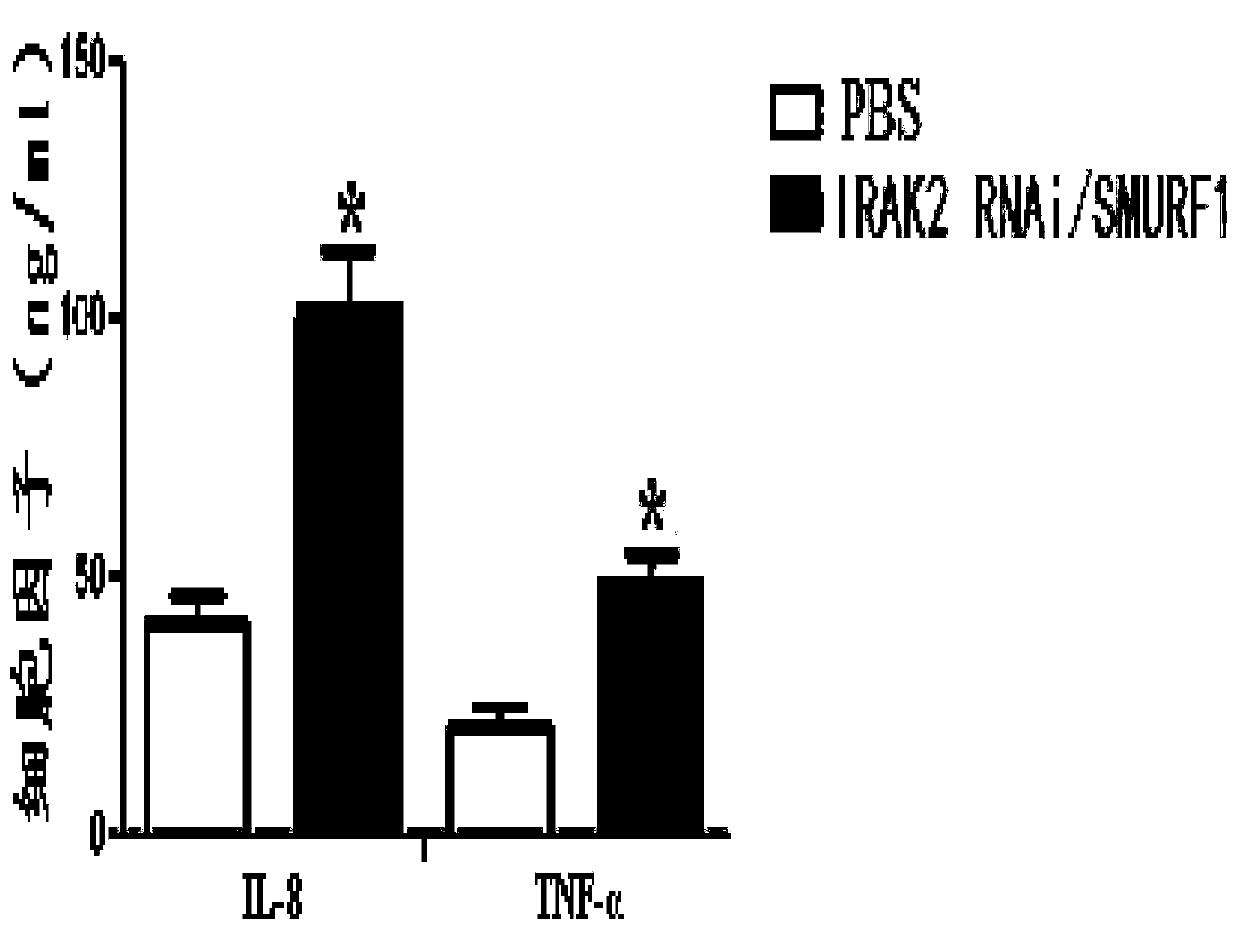
[0069] 25 μl of autologous whole blood was injected into the basal ganglia of mice, and 10 minutes later, 1 mg of double-gene recombinant complex macrophage vaccine or control (PBS) was injected at the same site. After 3 days, the mice were sacrificed, the brain tissues were obtained, and the cerebral edema of the mice was detected by dry and wet weight methods. Research results such as Figure 4 As shown, compared with the control (PBS), the double-gene recombinant complex macrophage vaccine can significantly reduce brain edema in mice with cerebral hemorrhage.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com