pFC330-BEC plasmid capable of achieving base precise point mutation and application thereof
A pfc330-bec, point mutation technology, applied in the field of genetic engineering, to achieve the effect of broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1. Construction of pFC330-BEC recombinant plasmid (pFC330-BEC plasmid) by gene recombination
[0035] The primer sequences used in Example 1 are shown in Table 1 (synthesized at Guangzhou Tianyi Huiyuan Biotechnology Co., Ltd.):
[0036] Table 1
[0037]
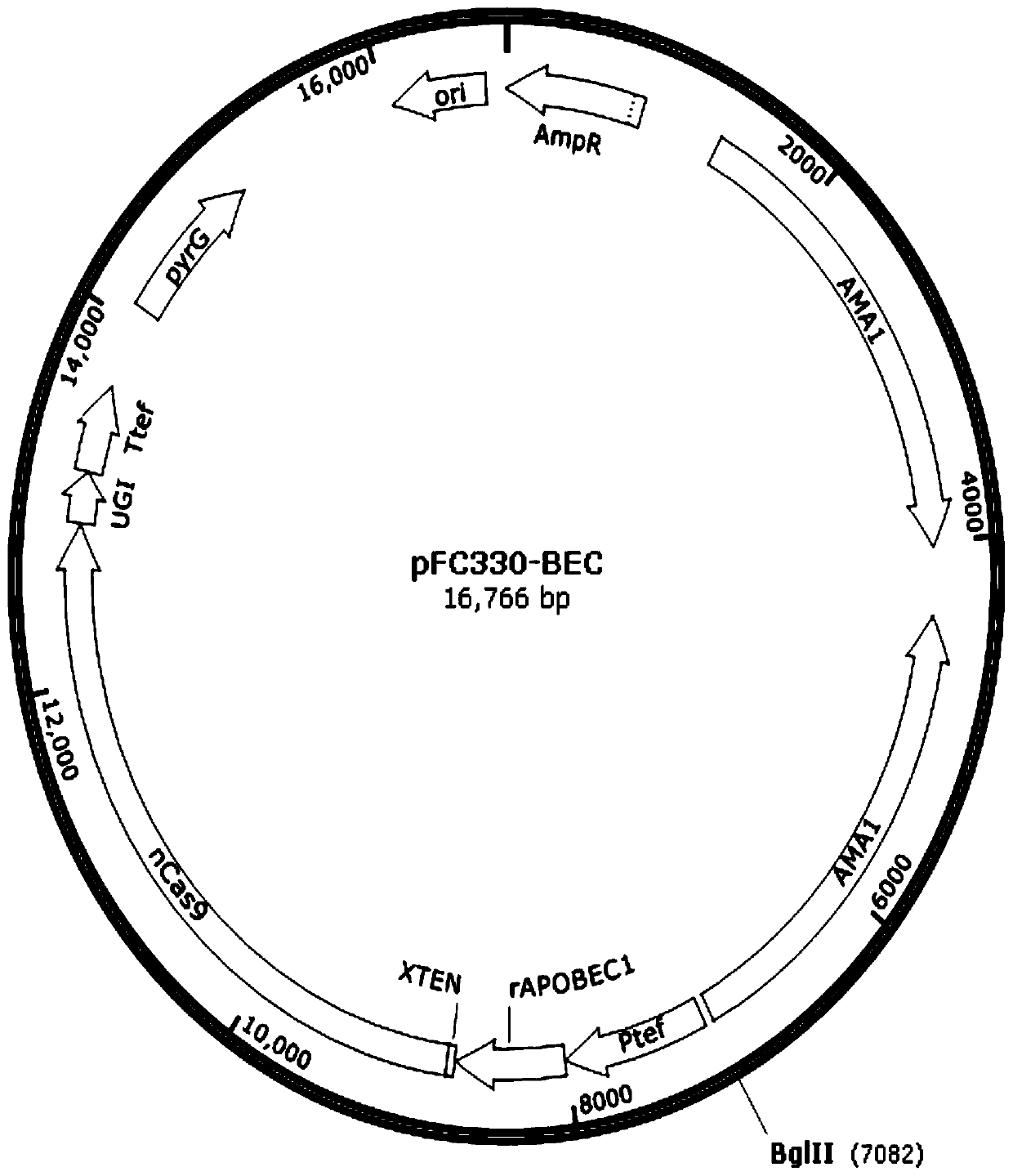
[0038] The composition of pFC330-BEC recombinant plasmid is as follows figure 1 As shown, among them, BglII site: for inserting sgRNA fragment; Ptef: Aspergillus fumigatus derived tef1 gene promoter, used to drive fusion protein expression; AmpR: ampicillin resistance gene; ori: plasmid replicator in Escherichia coli ( origin of replication); pyrG: uracil auxotrophic marker; AMA1: filamentous fungal autonomous replication sequence; rAPOBEC1: codon-optimized cytosine deaminase; nCas9: Cas9 protein with amino acid mutation at position 10 (D10A); UGI: Uracil DNA glycosylase inhibitory protein.
[0039] The pFC330-BEC recombinant plasmid contains an expressible rAPOBEC1-nCas9-UGI gene fragment and a filament...
Embodiment 2
[0045] Example 2. The pFC330-BEC plasmid achieves efficient base editing in Aspergillus niger CBS513.88 strain:
[0046] The pFC330-BEC plasmid can be used to achieve efficient base editing of different genes in Aspergillus niger CBS513.88 strain. In the experiment, the prtT gene (protease regulatory gene prtT) of Aspergillus niger was selected as an example, and the base editing experiment was carried out in the Aspergillus niger CBS513.88 strain.
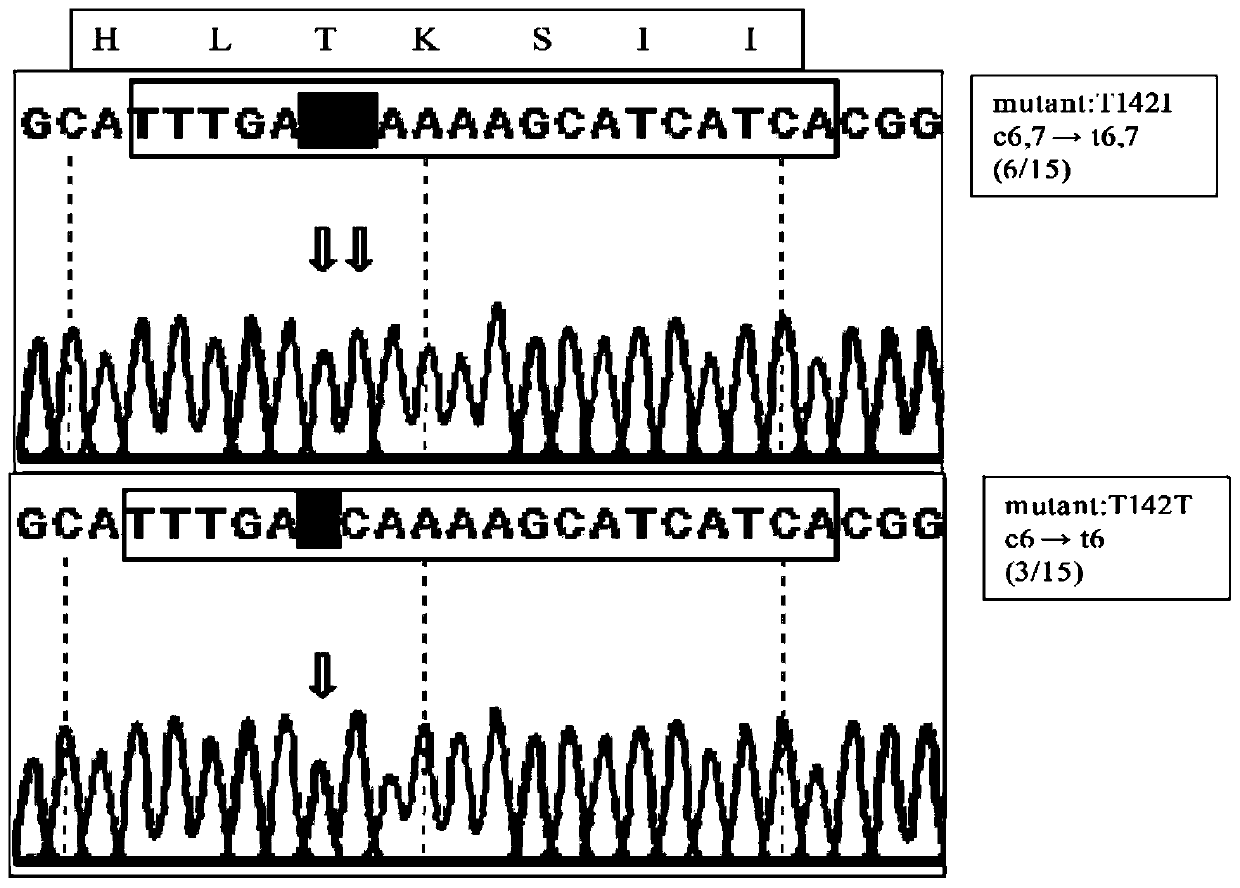
[0047] figure 2 It is a diagram of the sequencing results of base editing of the protease regulatory gene prtT in Aspergillus niger CBS513.88 strain using the pFC330-BEC plasmid in Example 2 of the present invention.
[0048] in figure 2 The DNA sequence (TTTGATTAAAAGCATCATCA) in the box is the protospacer sequence (20bp); the box above the sequencing map is the corresponding amino acid; the arrow indicates the mutation site; the right box is the amino acid position of the mutation and the corresponding transformant number. ...
Embodiment 3
[0073] Example 3. The pFC330-BEC plasmid achieves efficient base editing in Aspergillus niger FGSC1279 strain:
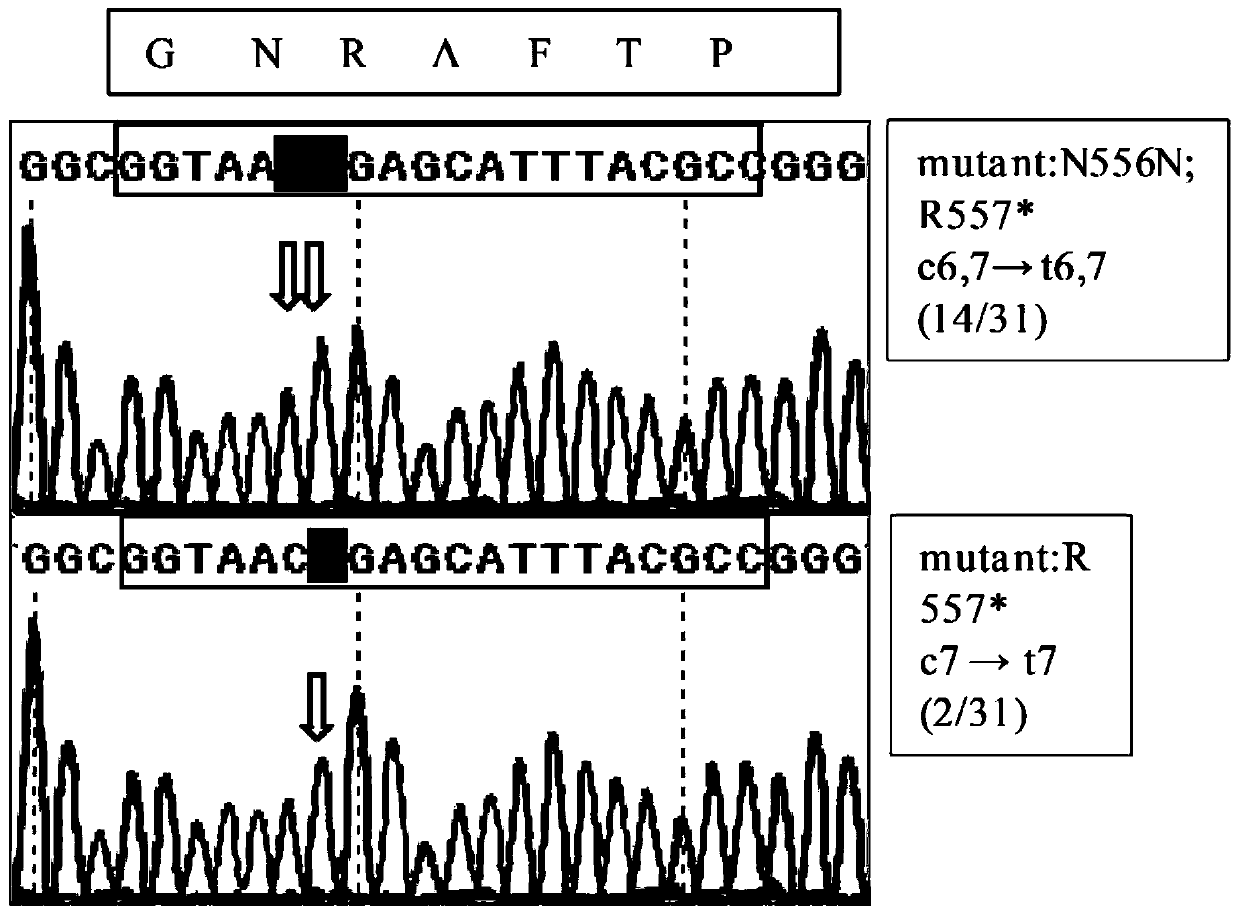
[0074] High-efficiency base editing of different genes can be achieved in Aspergillus niger FGSC1279 strain with pFC330-BEC plasmid. In the experiment, the pigment gene fwnA gene was selected as an example, and the base editing experiment was carried out in the Aspergillus niger FGSC1279 strain (see results in image 3 and Figure 4 ). image 3 The base mutation sequencing results of the fwnA gene in the Aspergillus niger FGSC1279 strain for the pFC330-BEC plasmid;
[0075] in image 3 The DNA sequence (GGTAATTGAGCATTTACGCC) in the box is the protospacer sequence (20bp); the box above the sequencing map is the corresponding amino acid; the arrow indicates the mutation site; the right box is the amino acid position of the mutation and the corresponding transformant number.
[0076] The arginine at position 557 was mutated into a stop codon by mutating the C at p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com