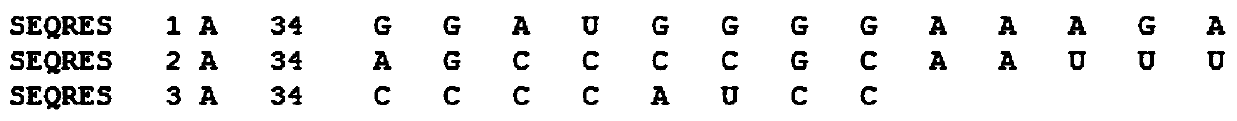Predicting method of RNA two-grade structure
A technology of secondary structure and prediction method, applied in the field of biological research, can solve the problems of difficult to obtain crystals, high cost, time-consuming secondary structure, etc., and achieve the effect of improving prediction efficiency and accurate prediction results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
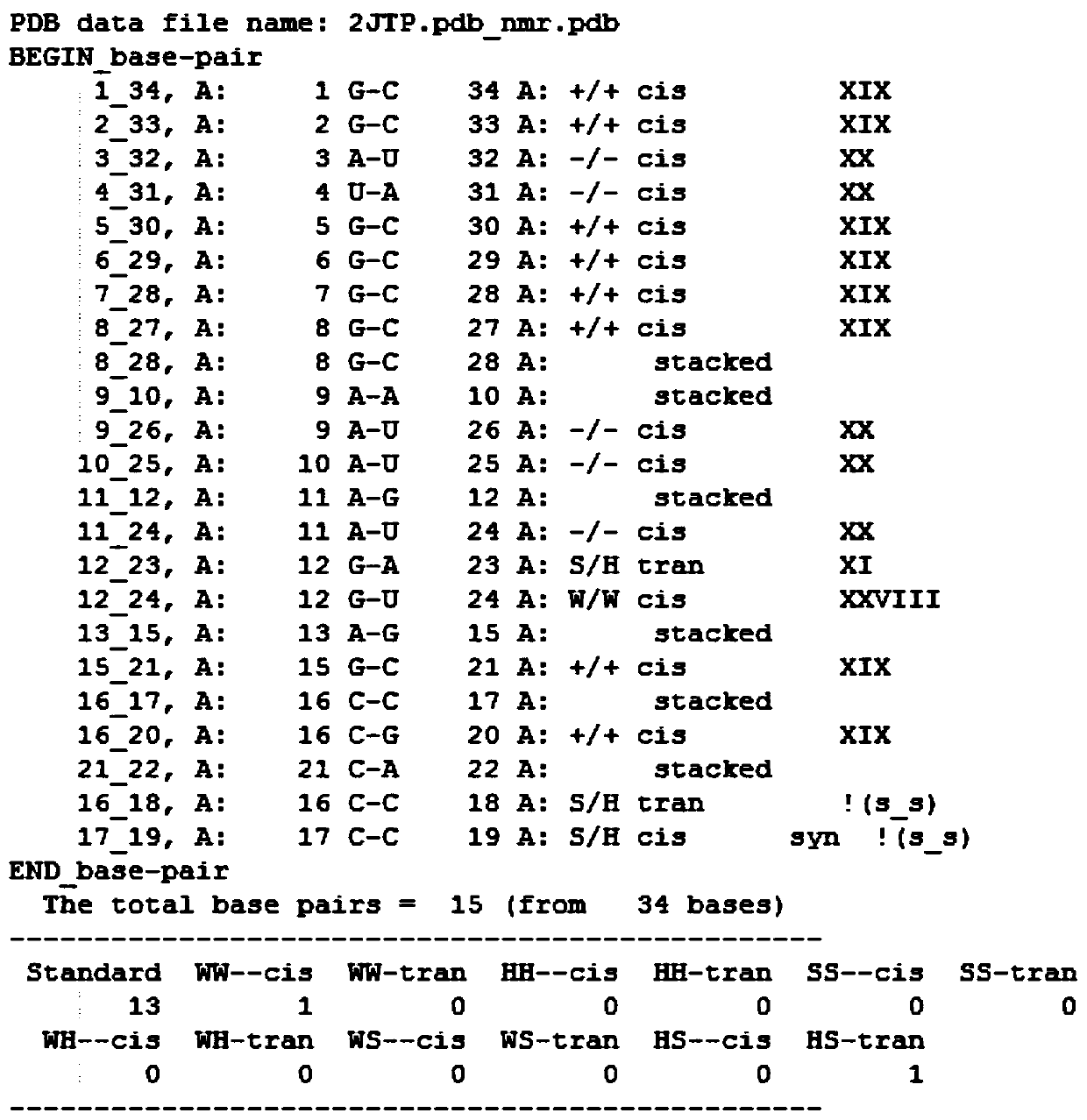
[0035] Example 1: A method for predicting RNA secondary structure: download the PDB data set from the official website of the PDB database, such as figure 1 Shown is a partial example of one of the RNA information '2JTP.pdb' in the downloaded PDB data. The PDB database contains three parts: RNA sequence information, RNA primary sequence and three-dimensional space coordinates. It can be seen from the figure The primary sequence of RNA is recorded in 'SEQRES'. First, data preprocessing is performed on the PDB data set, and the primary sequence is extracted by means of regular expressions. Some of the data are except A, C, G, U For other characters, these characters need to be cleaned to obtain the correct RNA primary sequence. Use the known RNA secondary structure prediction software RNAview to predict the RNA secondary structure corresponding to each primary sequence in batches under the Linux system, and remove the RNA tertiary structure with too high dimensionality, leaving ...
Embodiment 2
[0045] Example 2: A method for predicting the secondary structure of RNA: download the PDB data set from the official website of the PDB database, first perform data preprocessing on the PDB data set, and extract the primary sequence by means of regular expressions, some of which The data has characters other than A, C, G, and U. At this time, these characters need to be cleaned to obtain the correct RNA primary sequence. Use the known RNA secondary structure prediction software RNAview to predict the RNA secondary structure corresponding to each primary sequence in batches under the Linux system, and remove the RNA tertiary structure with too high dimensionality, leaving only the secondary structure and Partial pseudoknot structure. After data preprocessing, the PDB data set is divided into RNA primary sequence data set and RNA secondary structure data set, and then the RNA primary sequence in the RNA primary sequence data set is processed by computer, and a 5bit orthogonal 0...
Embodiment 3
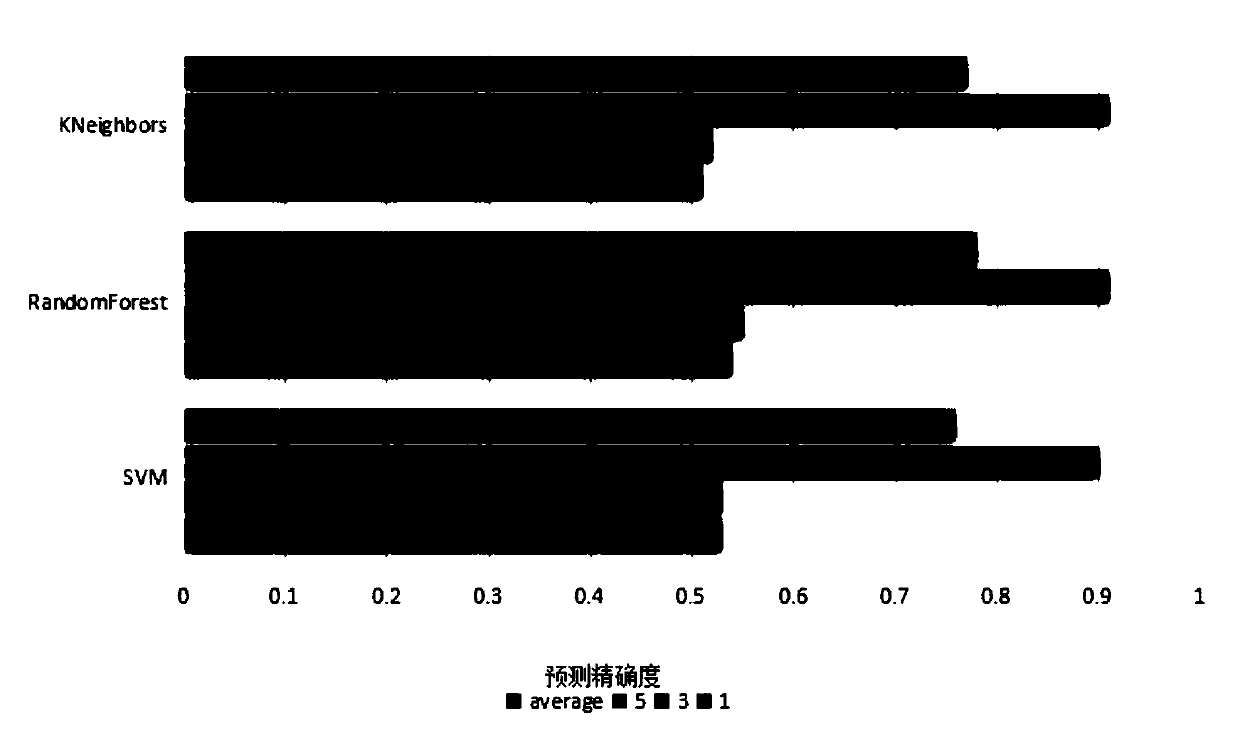
[0053] Embodiment 3: A kind of prediction method of RNA secondary structure, carry out the prediction of RNA secondary structure according to the operation step of embodiment 2, but when the RNA primary sequence after encoding is input into machine learning model as feature, adjust The number of windows and the number of base pairs in RNA long-range correlations were used to test the overall prediction accuracy of RNA secondary structure. In this embodiment, the SVM classifier is used alone, and the quantitative analysis method is used to determine the most suitable RNA secondary structure prediction. The test results are as follows: Figure 8 shown. From Figure 8 It can be seen that when there is no base pairing in the RNA long-range correlation, that is, when base pair=0, the more the number of windows, the highest overall prediction accuracy can reach 80%, because the base in the RNA secondary structure The relationship between becomes larger, the larger the number of wi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com