Protein coding method and protein post-translational modification site prediction method and system
A technology of post-translational modification and coding method is applied in the field of protein coding method and protein post-translational modification site prediction method and system, which can solve the problems of inability to predict protein post-translational modification with high precision, and inability to realize multi-feature data prediction.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
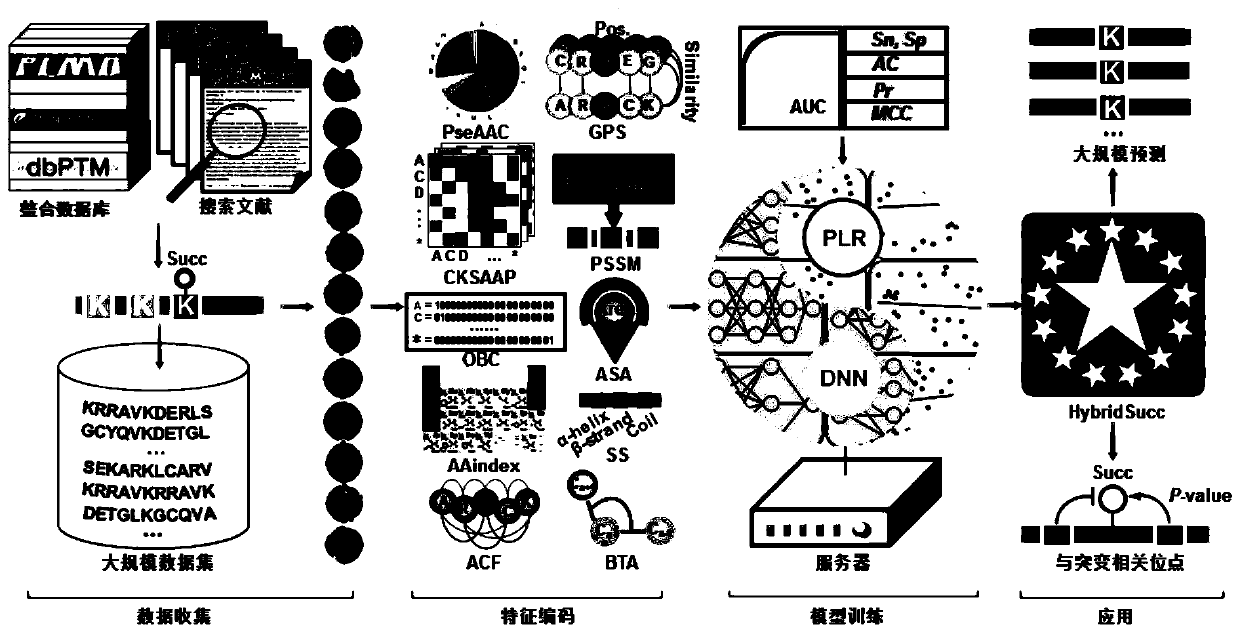
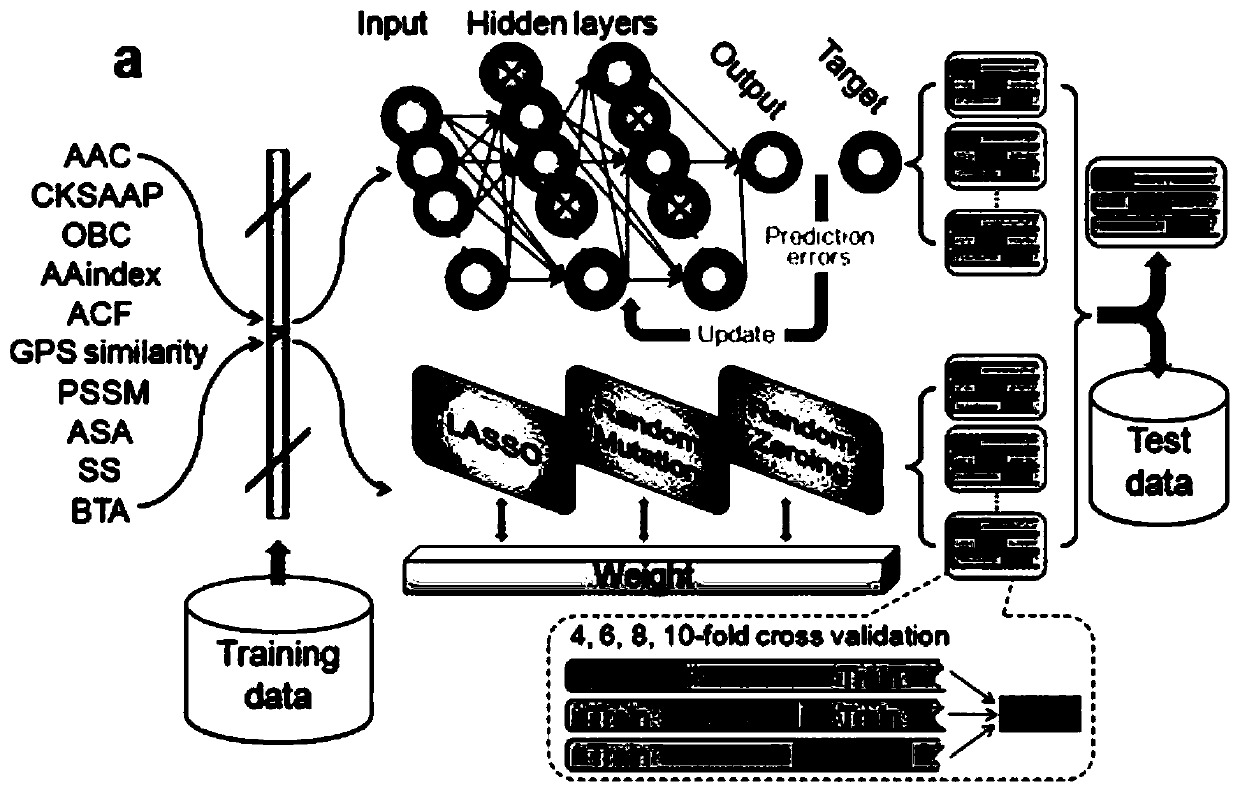
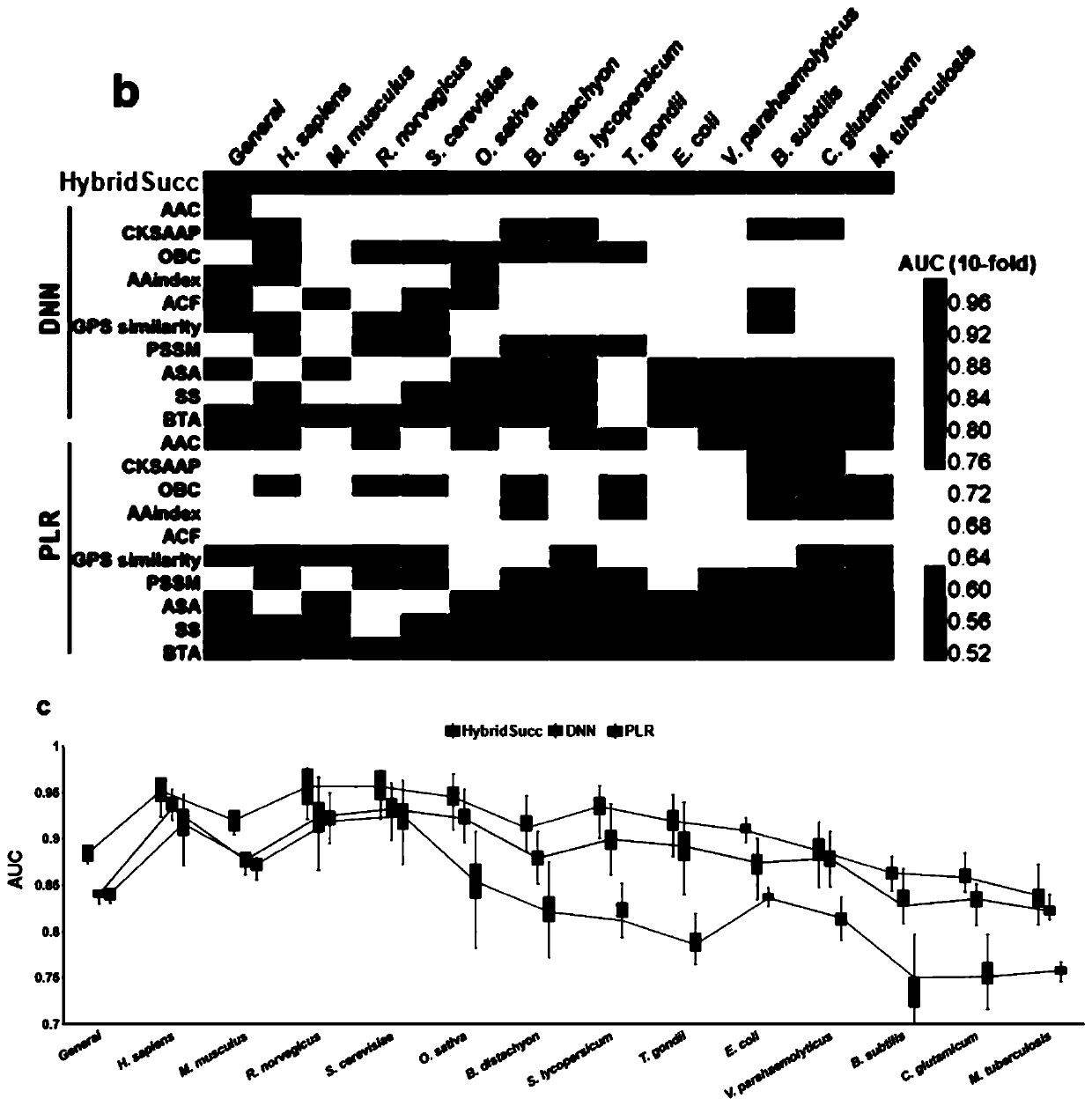
[0083] Taking protein lysine succinylation as an example, we use the method of the present invention to construct a prediction model named HybridSucc, and its flow chart is as follows figure 1 shown. The specific steps are:
[0084] 1. We collected and integrated 21,770 succinylation sites of 7,415 proteins from scientific literature, and downloaded the primary sequence of the protein from the UniProt database. Identified lysine succinylation sites were considered positive data, while remaining lysine sites in the same protein were considered negative data and classified species-specifically, as described for these succinylation sites The protein classifies loci into 13 species including human, mouse, yeast, rice, rat, E. coli. Cut the protein sequence into a sequence centered on the site, 10 amino acids upstream, 10 amino acids downstream, and a length of 21.
[0085] 2. Encode the protein sequence for features. Based on the data set, encode the positive and negative data ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com