Set of base editing artificial system for paddy
An artificial system and base editing technology, applied in the field of base editing artificial systems, can solve the problem that the efficiency is only one thousandth to two percent, and achieve the effect of high base editing efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] 1.1 Construction of pUbi:rBE14 and pUbi:rBE17
[0053] First, artificially synthesize the fragment of SEQ ID No.2, and then clone it into the cloning vector pUC57 (the vector is commercially available) to obtain pUC57-2. The amino acid sequence encoded by SEQ ID No.2 is shown in SEQ ID No.1. The segments of SEQ ID No.10 were artificially synthesized, and then cloned into the cloning vector pUC57 to obtain pUC57-10. The amino acid sequence encoded by SEQ ID No.10 is shown in SEQ ID No.11.
[0054] Then, clone SEQ ID No.6 (the ubiquitin promoter UbiP of maize), SEQ ID No.2, and SEQ ID No.8 (Nos terminator) into the pCAMBIA 1300 vector according to the direction from 5' to 3', and named is pUbi:rBE14. Cloning SEQ ID No.6 (maize ubiquitin promoter UbiP), SEQ ID No.10, and SEQ ID No.8 (Nos terminator) into the pCAMBIA 1300 vector according to the direction from 5' to 3', named pUbi: rBE17.
[0055] 1.2 Construction of pENTR4: sgRNA
[0056] According to the direction f...
Embodiment 2
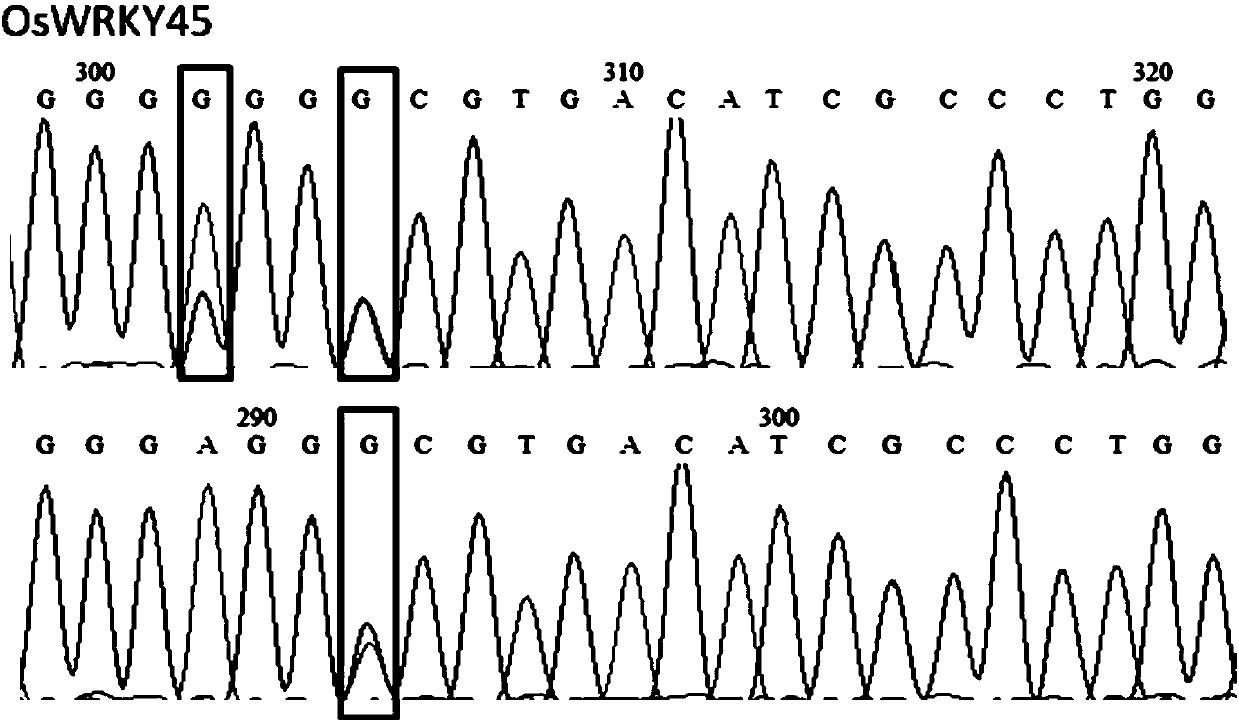
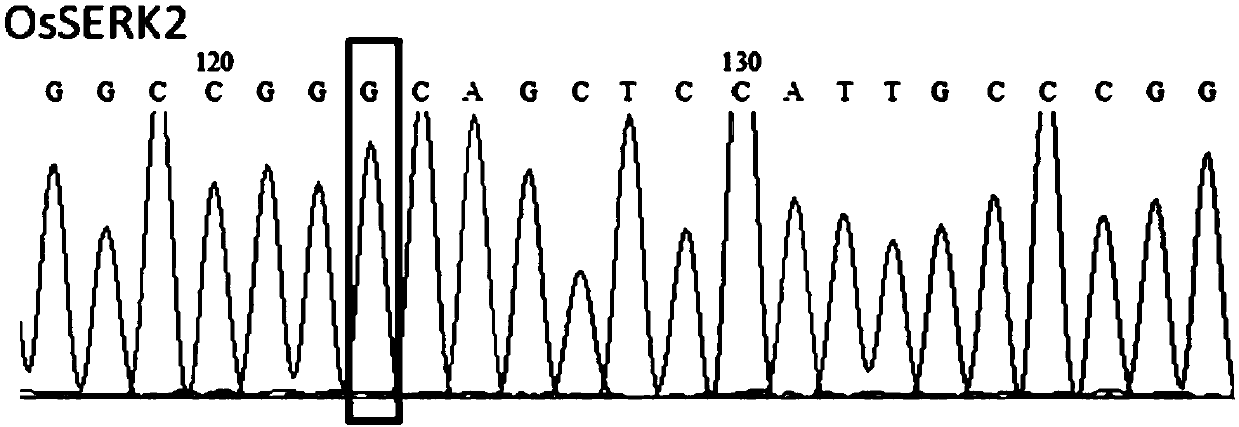
[0058] Base editing of OsWRKY45 and OsSERK2 using pUbi:rBE14
[0059] 2.1 Design and cloning of recognition sequences for OsWRKY45 and OsSERK2 genes
[0060] The transcript and genome sequences of each gene were obtained from the MSU / TIGR rice genome database (http: / / rice.plantbiology.msu.edu / ). Principles for designing target sequences: 1) Determine whether the editing site contains nucleotide base A or T, and whether the amino acid changes caused by the mutation of the corresponding base A to G or the corresponding base T to C are in line with expectations; When the nucleotide sequence of T is used as the target nucleotide sequence, its reverse complementary sequence is used for base editing; 2) Search for the PAM motif sequence in the 3' end direction, so that the mutant base A or T is in the target nucleus Positions 4 to 7 in the direction of the 5' end of the nucleotide sequence. Wherein, the PAM motifs recognized by the pUbi:rBE14 system are NGG, NAG, GAGN and AAGN, et...
Embodiment 3
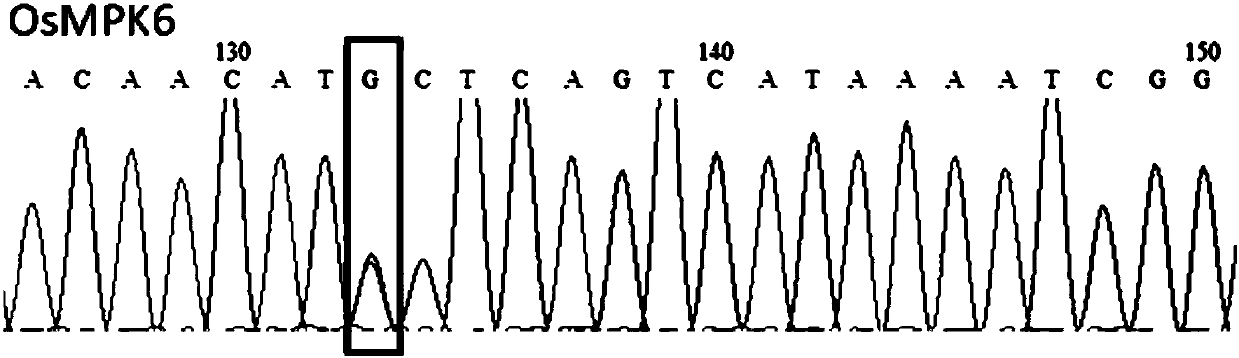
[0081] Base editing of OsMPK6 using pUbi:rBE14 and pUbi:rBE17, respectively
[0082] The PAM motifs recognized by the pUbi:rBE14 system and the pUbi:rBE17 system are NGG, NAG, GAGN and AAGN, etc., and the N is A or G or C or T.
[0083] According to the target nucleotide sequence primer design principle in embodiment 2, the target nucleotide sequence of OsMPK6 is designed to contain the primer that matches with the BsaI restriction site end connection: gOsMPK6-F1 (SEQ ID No.22) and gOsMPK6-R1 ( SEQ ID No. 23).
[0084] After synthesizing the primers, use T4 polynucleotide kinase to phosphorylate the primers, anneal to form double strands, clone gOsMPK6-F1 / R1 into the BsaI restriction site of the pENTR4:sgRNA vector, and confirm that the inserted fragment is completely correct by sequencing . Wherein, the forward primer represents the target nucleotide sequence inserted into the BsaI site on the pENTR4:sgRNA carrier, as shown in SEQIDNo.24. Wherein, the 2nd, 4th and 7th posi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com