Saccharum spontaneum retrotransposon sequence and identification method thereof
A technology of retrotransposon and identification method, which is applied in the field of retrotransposon sequence and its identification, which can solve the problem of small shape, poor effect, and lack of rapid and accurate identification of the chromosome Probe and other problems, to achieve the effect of good primer specificity, single and bright band
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
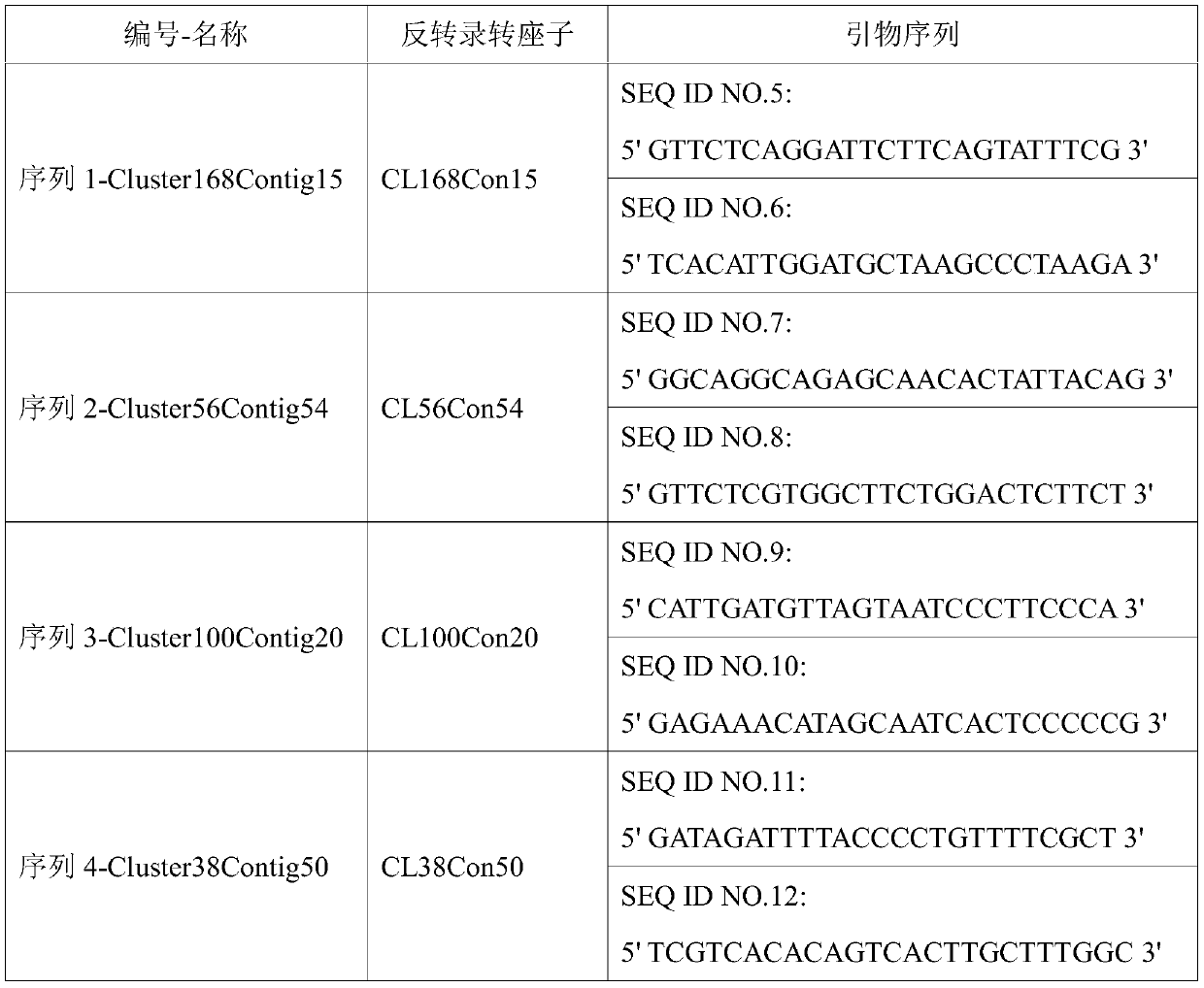
[0035] Example 1: According to the genome clustering data analysis of SES208 and tropical species LA Purple, the Sequence Retrotransposon sequence of Sequence 1-Cluster168Contig15, Sequence 2-Cluster56Contig54, Sequence 3-Cluster100Contig20, Sequence 4-Cluster38Contig50, the nucleotide sequences of which are respectively shown in SEQ ID NO.1-4. The molecular marker primer sequences corresponding to the sequence 1-Cluster168Contig15, the sequence 2-Cluster56Contig54, the sequence 3-Cluster100Contig20, and the sequence 4-Cluster38Contig50 are shown in SEQ ID NO.5-12.
[0036] Table 1 Amplification primers for reverse transcription of transposon sequence
[0037]
[0038] Primers were designed for retrotransposon sequences using Primer Premier 5.0.
[0039] Perform PCR amplification on the above four cut-hand-closed retrotransposon sequences. The PCR amplification conditions are: pre-denaturation at 95°C for 3 minutes; denaturation at 98°C for 30 seconds, annealing and extens...
Embodiment 2
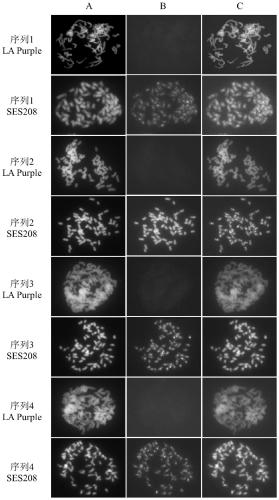
[0040] Example 2: Identification by fluorescence in situ hybridization (FISH)
[0041]Fluorescence in situ hybridization is a technique for detecting the distribution of target sequences on chromosomes. The detection results of this method are true and reliable. It is not only suitable for the identification of single-copy genes on chromosomes, but also for the identification of multiple copies of repetitive sequences on chromosomes. Identification studies.
[0042] The fluorescence in situ hybridization identification of the cutshoumi retrotransposon sequence includes the following steps:
[0043] (1) Take the vigorously growing SES208 and LA Purple root tips respectively, and put them into 8-hydroxyquinoline solution at room temperature for pretreatment;
[0044] (2) cutting off the root cap and the elongation zone, leaving the root tip meristem, and using cellulase and pectinase to enzymatically remove the root tip meristem cells at 37°C;
[0045] (3) Aspirate the root ti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com