Parsing of biological core genome information through low-cost assembling based on apparent group information
A genome and genome sequence technology, applied in the field of biological assembly analysis, can solve the problems of underestimated polymorphism, high requirements for genome sequence information, and difficulty in meeting the genetic analysis of large genome species populations.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
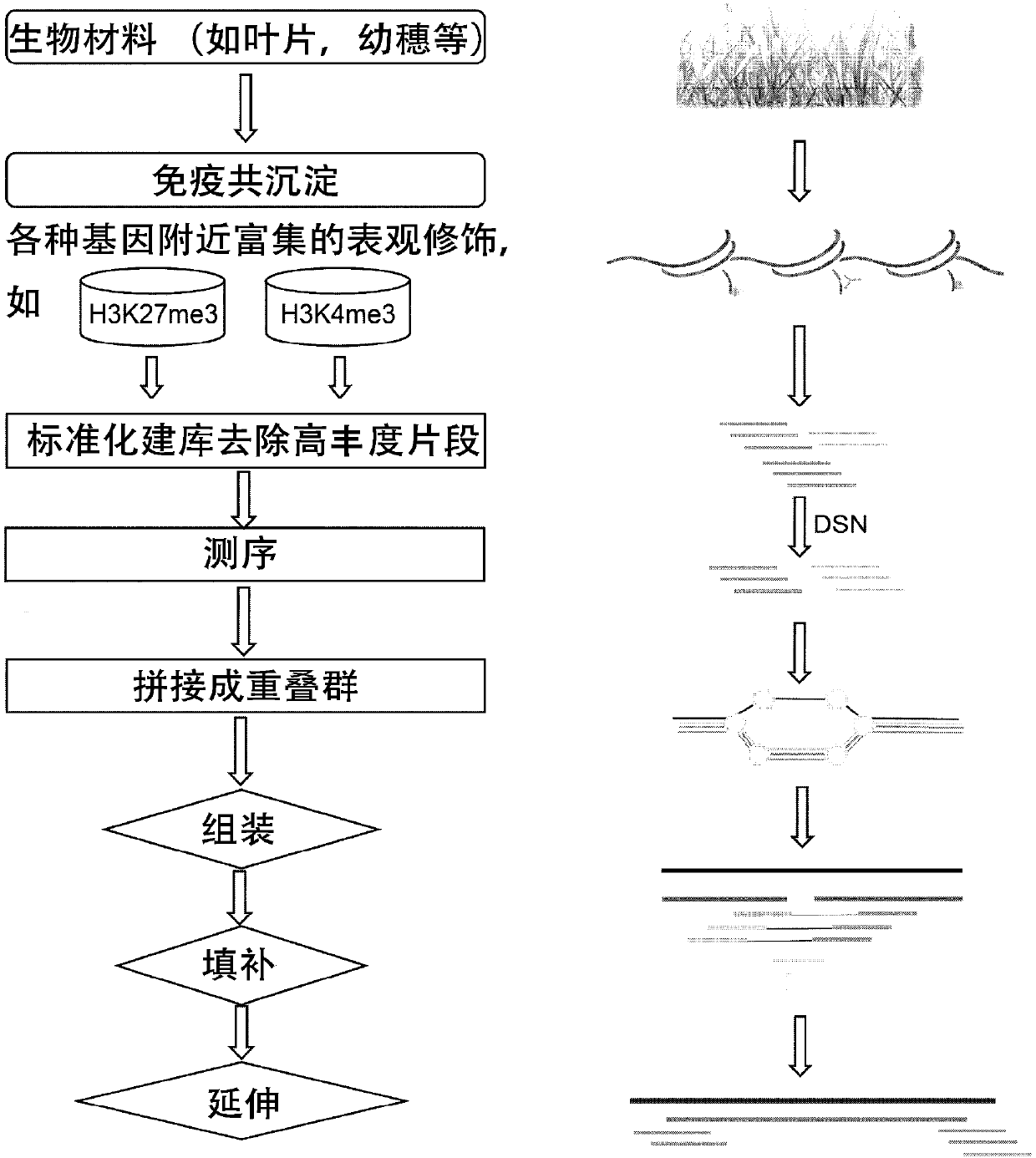
[0073] Embodiment 1, genetic polymorphism experimental method and calculation process of the present invention
[0074] 1. Chromatin immunoprecipitation (ChIP)
[0075] 1. Select the materials of the corresponding organizations in the required period.
[0076] 2. Put the materials in pre-cooled Fix Buffer on ice (use 100ml Fix Buffer for each material). If the material is too large, the material can be cut into pieces.
[0077] 3. Vacuum pumping (10min), cover the mouth of the bottle with gauze to prevent the material from spraying out during pumping.
[0078]4. Add 5ml of 2.5M glycine and continue pumping for 5 minutes.
[0079] 5. Wash 5 times with deionized water, the last time remove as much excess water as possible.
[0080] 6. Fully grind the material with liquid nitrogen, and grind at least 4 times.
[0081] 7. Pour the powder into a 50ml Falcon tube and quantify. The process was performed in liquid nitrogen. Can be stored at -80°C.
[0082] 8. Add 15ml of CLB1 ...
Embodiment 2
[0127] Embodiment 2, the assay analysis of wheat
[0128] In this example, taking Chinese spring wheat as an example, the important evaluation parameters after the simplified genome sequencing scheme are analyzed in sequence.
[0129]Chinese spring wheat was obtained, and H3K4me3 and H3K27me3 antibodies were used to perform co-immunoprecipitation using the method described in Example 1 above. Afterwards, ChIP-seq library construction and sequencing were performed. Then perform CGT-Seq de novo assembly. After that, the steps of assembling, filling and extending are carried out.
[0130] The effect evaluation is as follows:
[0131] 1. Accuracy
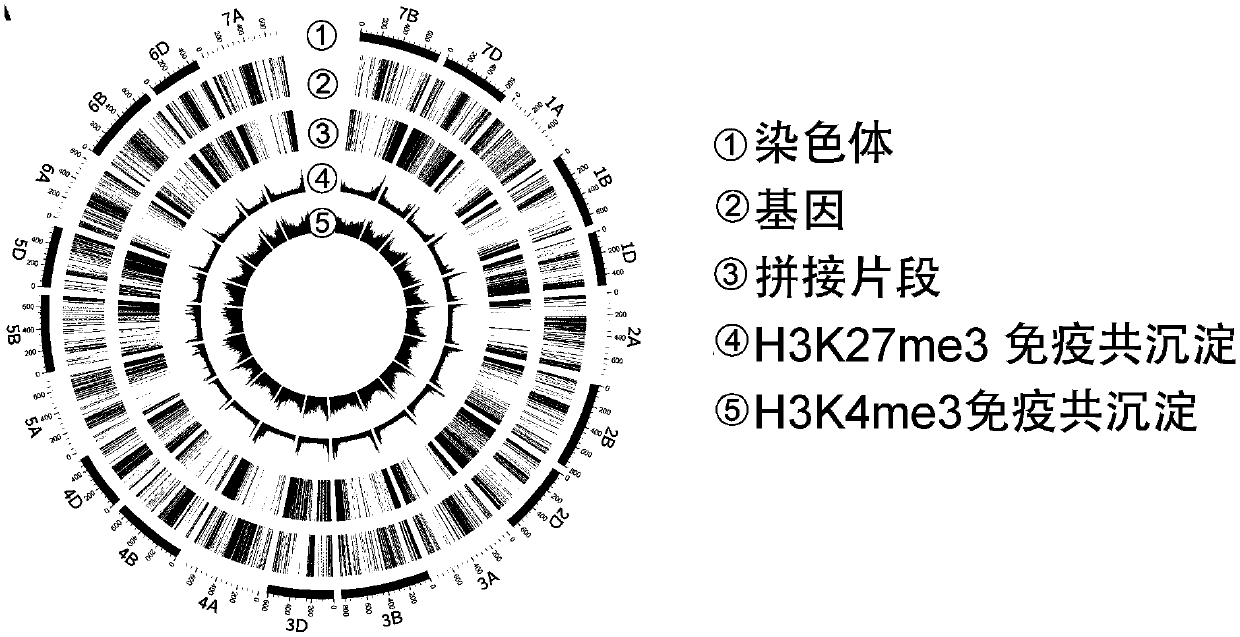
[0132] Using Circos (http: / / circos.ca / ) mapping software, compare the sequence obtained by using the method in Example 1 above with the sequenced Chinese spring wheat data (the latest RefSeq version v1.0 published by the International Wheat Sequencing Consortium) , the result is as figure 2 As shown, the fragments captured by H3K...
Embodiment 3
[0144] Embodiment 3, the assay analysis of rice
[0145] Chinese spring wheat was obtained, and H3K4me3, H3K4me1, H3K27me3, H3K27ac, H3K36me3 were used as co-immunoprecipitation antibodies, and the co-immunoprecipitation was carried out by the method described in the foregoing Example 1. Afterwards, ChIP-seq library construction and sequencing were performed, followed by CGT-Seq de novo assembly.
[0146] The effect evaluation is as follows:
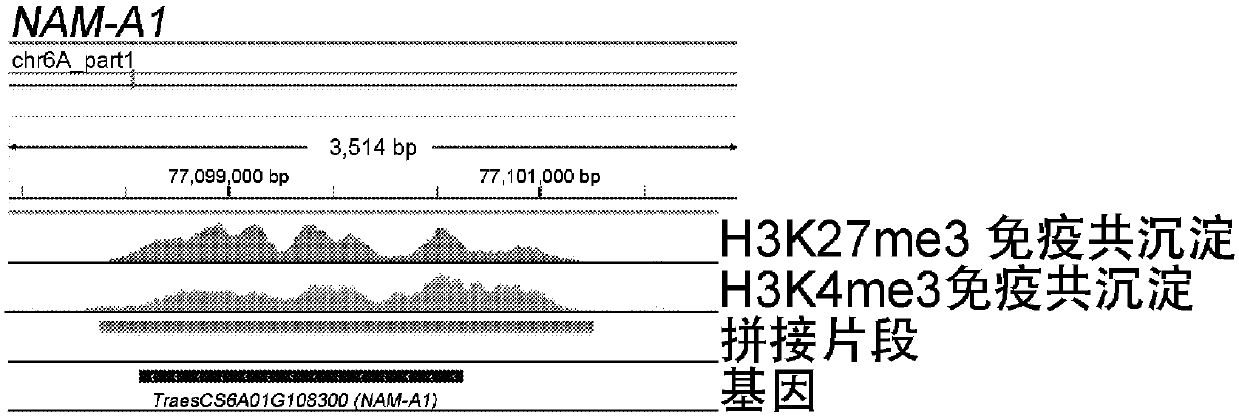
[0147] First, by comparing with gene position and gene expression ( Image 6 ), it can be seen that H3K4me3, H3K4me1, H3K27ac, and H3K36me3 modifications are positively correlated with gene expression, and are mainly distributed near highly expressed genes, among which H3K4me3 is most widely modified; H3K27me3 is negatively correlated with gene expression, mainly distributed in non-expressed and low-expressed genes. near expressed genes. Therefore in all subsequent experiments the combination of H3K4me3 and H3K27me3 was used for analy...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com