Serotyping method of Actinobacillus pleuropneumoniae, primer set and PCR (polymerase chain reaction) systems
A technique for porcine pleuropneumoniae and Actinobacillus, which is applied in the field of molecular biology, can solve the problems of high cost, small identification range of PCR typing method, long time consumption, etc., and achieves the effects of less reagents, large detection range and high efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] The establishment of embodiment 1 primer grouping system
[0048] In this embodiment, specific primers are designed for each serotype by comparing the conserved gene and the outer lipid membrane protein (OlmA) gene in the capsular synthetic gene cluster of Actinobacillus pleuropneumoniae of serotype 1-15; wherein, Serum type 9 and serotype 11 use the same characteristic gene, which makes the specific sequences of type 9 and type 11 the same, a total of 14 pairs of specific primers, the sequences of which are SEQ ID NO.1-SEQ ID NO.28. In order to make it difficult for the primers in the same PCR system to be reversely complementary to form dimers, and to easily distinguish the size of the amplicon, the specific primers are divided into 4 groups, as follows:
[0049] Table 1 Primer grouping system
[0050]
[0051]
Embodiment 2
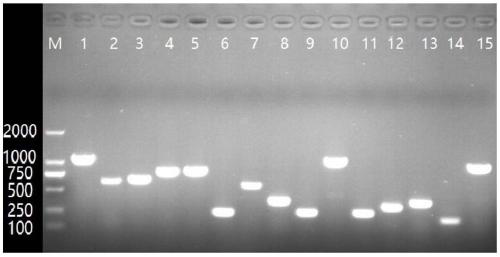
[0052] Embodiment 2 Each serotype primer specificity experiment of Actinobacillus pleuropneumoniae strain
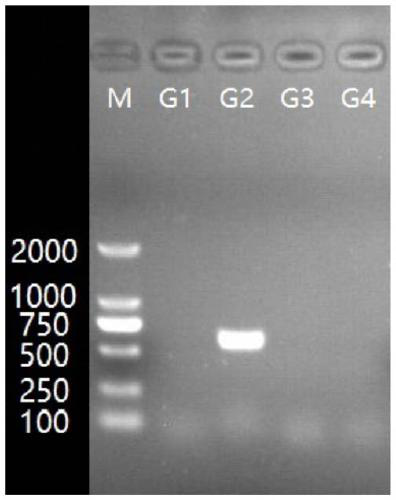
[0053] In this embodiment, each serotype-specific band is obtained by the following method:
[0054] 1) Streak the Actinobacillus pleuropneumoniae strains of serotype 1-15 on the TSA medium plate respectively, cultivate a single colony, pick a single colony, boil in a water bath, centrifuge, and obtain the genomic DNA of the strain template;
[0055] 2) Using the genomic DNA obtained in step 1) as a template, respectively using the corresponding specific primers to carry out PCR amplification, the amplification conditions are as follows:
[0056] 10 μL reaction system: 5 μL of 2×Premix Taq, 0.4 μL of each 10 mmol / mL primer, 0.5 μL of template DNA, and then add sterile double distilled water or ultrapure water to make up to 10 μL;
[0057] PCR reaction program: pre-denaturation at 95°C for 5 minutes; then denaturation at 95°C for 30 seconds, annealing at 57°C for 30 sec...
Embodiment 3
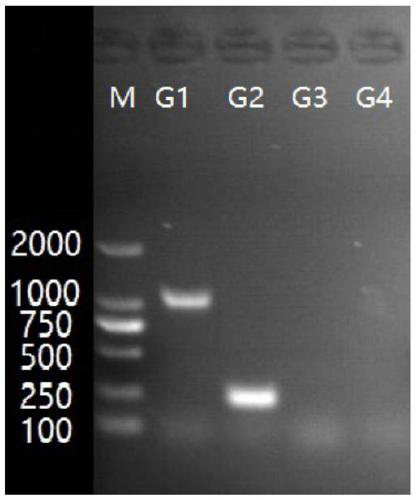
[0060] The multiple PCR serotyping method of embodiment 3 Actinobacillus pleuropneumoniae bacterial strain
[0061] The present embodiment carries out the serotyping of Actinobacillus pleuropneumoniae bacterial strain by following method:
[0062] 1) Streak the Actinobacillus pleuropneumoniae strains of serotype 1-15 on the TSA medium plate respectively, cultivate a single colony, pick a single colony, boil in a water bath, centrifuge, and obtain the genomic DNA of the strain template;
[0063] 2) Using the genomic DNA obtained in step 1) as a template, respectively use the 4 sets of primers provided in Example 1 to carry out PCR amplification, and the amplification conditions are as follows:
[0064] 10 μL reaction system: 5 μL of 2×Premix Taq, 0.4 μL of each 10 mmol / mL primer, 0.5 μL of template DNA, and then add sterile double distilled water or ultrapure water to make up to 10 μL;
[0065] PCR reaction program: pre-denaturation at 95°C for 5 minutes; then denaturation at...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com