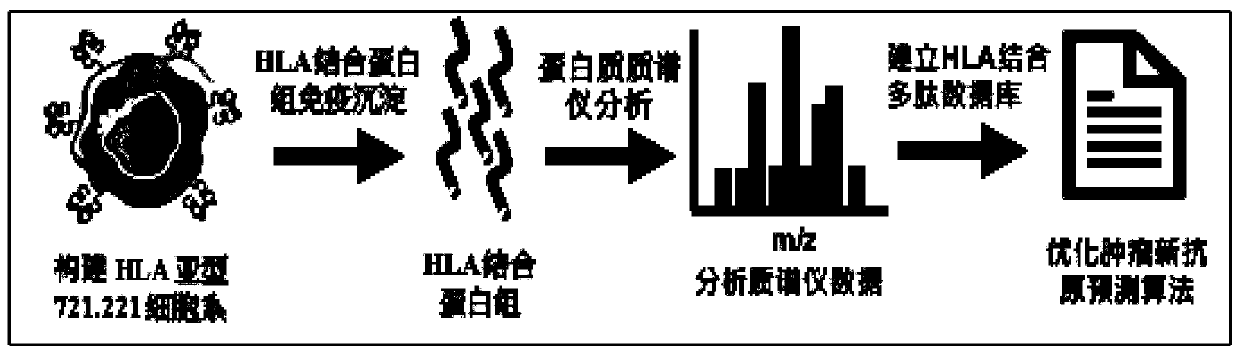
Tumor neoantigen prediction platform and application thereof in neoantigen vaccine development system
A platform and new technology, applied in the fields of molecular biology and bioinformatics, can solve problems such as low accuracy, and achieve the effect of improving prediction accuracy, screening and R&D efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1: Construction of HLA subtype 721.221 cell line
[0032] 1. Using HLA locus-specific primers to copy HLA gene fragments from HLA homozygous human B lymphocyte cell lines
[0033] (1) Using the RNeasy kit to obtain HLA homozygous human B lymphocytes (cultured in RPMI cell culture medium, containing 10% fetal bovine serum, 1% penicillin / streptomycin mixed solution / glutamine and 1mM sodium pyruvate ) to extract total RNA;
[0034] (2) Add Oligo dT primers, dNTP mixture, dithiothreitol and MMLV reverse transcriptase to the extracted total RNA to produce cDNA;
[0035] (3) Add 2 μl cDNA to PCR buffer, 200 μM dNTP mixture, 0.3 μM HLA locus-specific primers and 0.5 μl Taq polymerase to carry out PCR reaction;
[0036] The operating procedures of the thermal cycler are shown in Table 1 below:
[0037] Table 1 Operation procedure of thermal cycler
[0038]
[0039] The sequences of HLA locus-specific primers are shown in Table 2 below:
[0040] Table 2 HLA locus...
Embodiment 2
[0063] Example 2: Using high-sensitivity protein mass spectrometry to analyze the binding protein groups of 55 HLA subtype MHC-negative 721.221 cell lines with a high proportion of the Chinese population and construct an HLA-binding polypeptide database
[0064] The present invention uses the HLA subtype gene frequency database (http: / / www.allelefrequencies.net) to screen out a total of 55 HLA-A and HLA-B subtypes with a high proportion in the Chinese population, covering a total of 98.47% of HLA-A subtypes and 89.39% of HLA-B subtypes in the Chinese population. The 55 HLA subtypes include 24 HLA-A subtypes and 31 HLA-B subtypes, the specific numbers are as follows: HLA-A*01:01, HLA-A*02:01, HLA-A*02:02 , HLA-A*02:03, HLA-A*02:04, HLA-A*02:05, HLA-A*02:06, HLA-A*02:07, HLA-A*03:01, HLA -A*11:01, HLA-A*11:02, HLA-A*23:01, HLA-A*24:02, HLA-A*24:03, HLA-A*26:01, HLA-A *29:02, HLA-A*30:01, HLA-A*31:01, HLA-A*32:01, HLA-A*33:01, HLA-A*33:03, HLA-A*66 :01, HLA-A*68:01, HLA-A*68:0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com