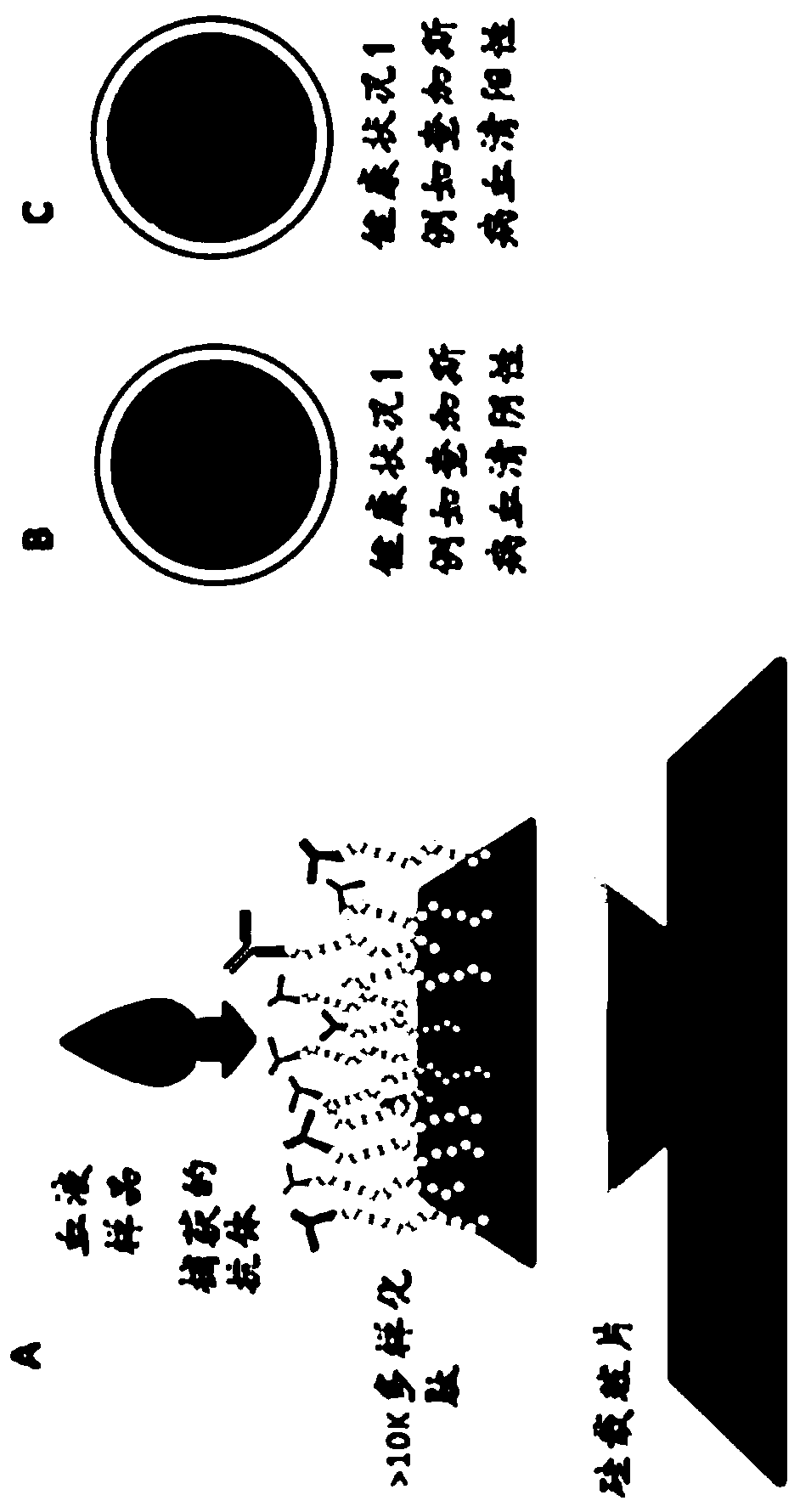
Methods for screening infections
A technology for trypanosoma cruzi infection and subjects, applied in the field of screening infection, which can solve the problems of intensive and complicated labor, and the inability of diagnostic tools to reliably detect the early stage of infection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 4
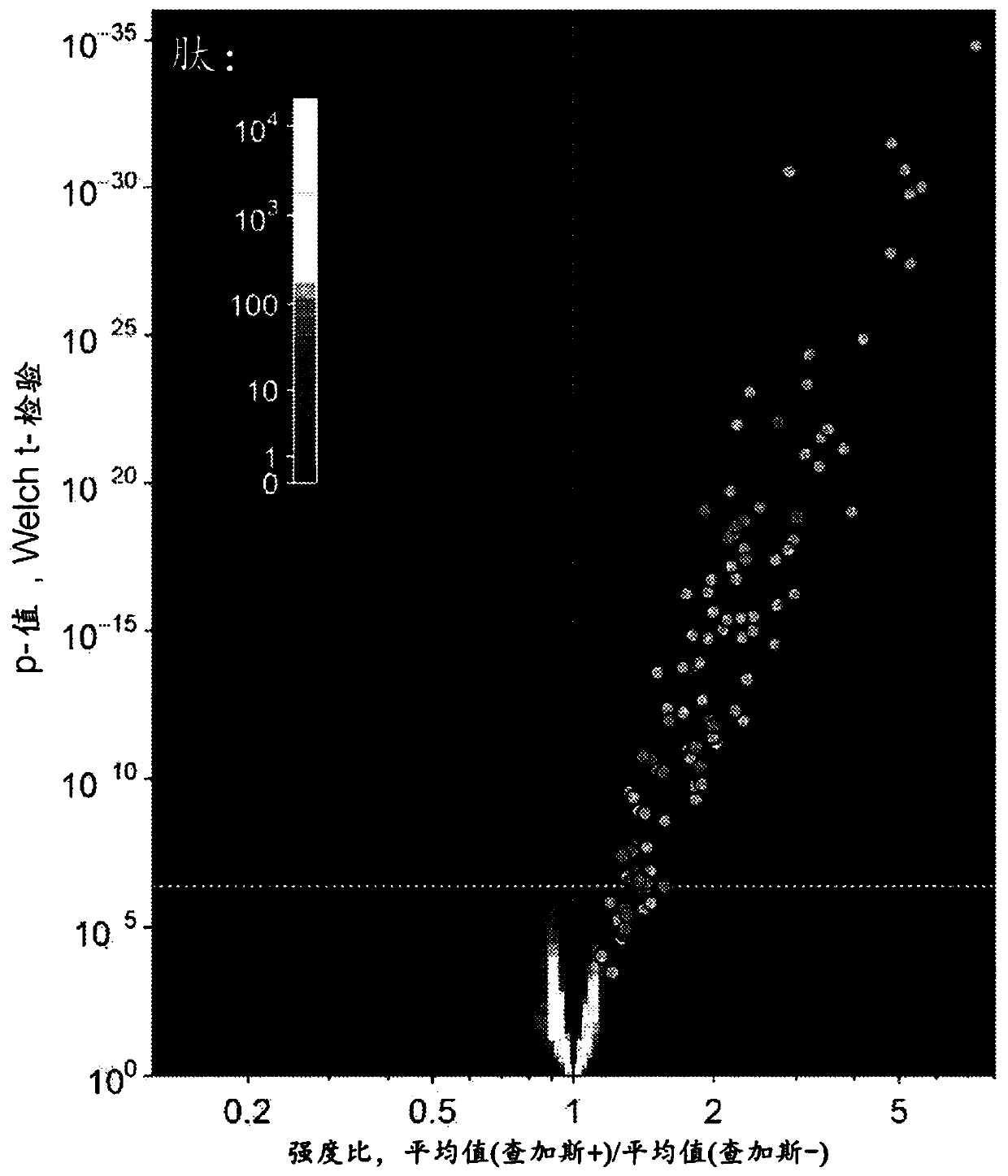
[0123] Example 4 illustrates a method for identifying candidate protein biomarkers using a discriminative peptide that differentiates serological status in samples from healthy subjects from those from subjects infected with Trypanosoma cruzi (Chagas disease). sample. A healthy subject may be a subject who was previously infected with Trypanosoma cruzi and seroconverted to seronegative, and / or a subject who has never been infected with Trypanosoma cruzi. Table 2 provides a list of candidate protein biomarkers. Similarly, candidate protein biomarkers can be identified using discriminative peptides that distinguish samples from subjects with other infectious diseases from samples from healthy subjects, samples from subjects with other infectious diseases and the serological status of samples from subjects with simulated disease (which may or may not be infectious).
[0124] In some embodiments, a method for identifying a candidate protein biomarker for an infectious disease co...
Embodiment 1
[0196] Example 1 - Immunosignature method for diagnosing infection
[0197] An immunosignature assay was developed to detect and differentiate T. cruzi, HBV, HCV, and WNV infections according to the following.
[0198] Donor samples. Plasma samples from donors seropositive for Chagas antibodies, as well as age- and sex-matched healthy donor plasma, and for hepatitis B virus (HBV), hepatitis C virus (HCV), or West Nile virus (WNV) Plasma samples that tested seropositive were obtained from Creative Testing Solutions (Tempe, AZ). Two sample cohorts were obtained, one in 2015 and the other in 2016. Upon receipt, plasma was thawed, mixed 1:1 with ethylene glycol as a cryoprotectant, and aliquoted into single-use volumes. Single-use aliquots were stored at -20°C until needed. The remaining sample volume was stored neat at -80 °C. The identity of all samples was tracked using tubes (Micronic, Leystad, the Netherlands) with two-dimensional barcodes. In preparation for the assay,...
Embodiment 2
[0220] Example 2 - Platform Verification
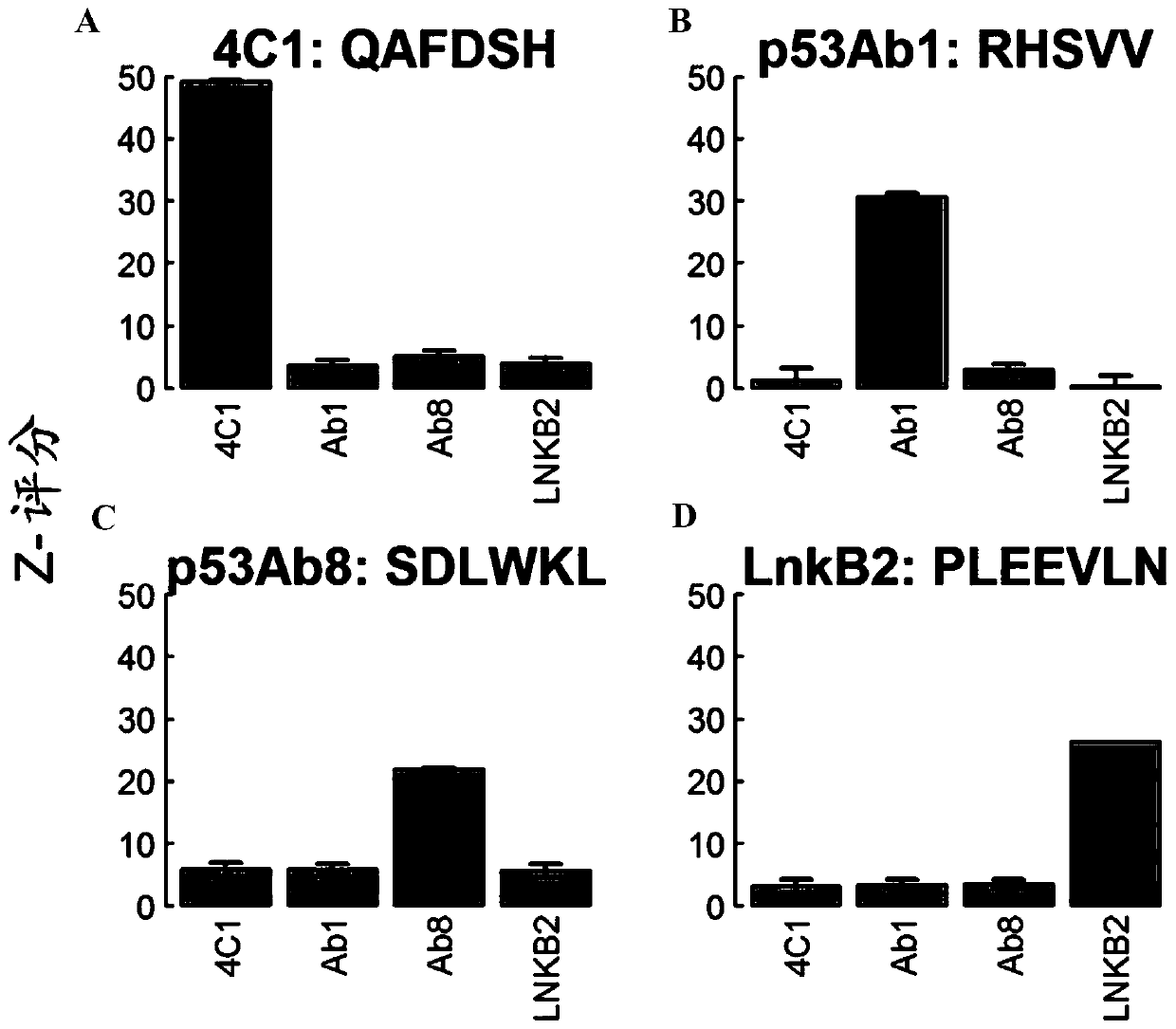
[0221] Experiments were performed using monoclonal antibodies to evaluate the quality of the final in situ synthesized array peptide products with respect to ligand presentation and antibody recognition.
[0222] All diagnostic assays are performed on a validated microarray platform.
[0223] A protocol for peptide synthesis was developed in which parallel coupled reactions were performed directly on silicon wafers using masking and photolithography. Antibody binding events were queried using arrays representing a total of 131,712 peptides (median length 9 amino acids) at each 14 μm x 14 μm feature. The array layout included 126,009 library peptide features and 6203 control peptide features attached to the surface via a common linker (see Example 1). The library peptides were designed to sample all possible amino acid combinations evenly. Control peptides included 500 features corresponding to established epitopes of five different...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com