Method for efficiently knocking chimeric antigen receptor (CAR) genes into T cell specific genome loci through clusters of regularly interspaced short palindromic repeats (CRISPR)-Cas9, and application of method
A genome and gene technology, applied in the field of cell therapy, can solve the problems of CAR-T cell exhaustion, more difficult, cancerous and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0098] Embodiment 1: PBMC extraction
[0099] Recruit healthy volunteers (inconvenience to disclose information), without symptoms of cold and fever. Professional medical personnel take 100ml of blood from the median vein of the human elbow and put it into the BD anticoagulant blood vessel. After blood collection, the blood was mixed with an equal amount of PBS buffer (containing 2% fetal bovine serum). Take the PBMC separation tube Sepmate-50, carefully add 15ml of Ficoll buffer, then add the blood PBS mixture, carefully add about 30ml to each tube. After centrifuging at 1200g for 10 minutes, quickly pour the supernatant into a new 50ml tube, centrifuge at 200g for 8 minutes, discard the supernatant, add 10ml PBS buffer to resuspend the pellet, discard the supernatant, add 10ml PBS buffer to resuspend, centrifuge and discard After clearing, resuspend all pellets in 10ml of supernatant PBS buffer. To count the resuspended cells, take 10 μl of the suspension and add 10 μl of...
Embodiment 2
[0102] Example 2: T cell activation
[0103] Take the above PBMC cells 1×10 7 One, resuspended in 6ml VIVO-15 medium, activated with anti-CD3 / anti-CD28 antibody magnetic beads. Take anti-CD3 / anti-CD28 antibody magnetic beads (Life Technology) 6×10 6 Each was resuspended with PBS buffer (containing 2 mM EDTA and 1% fetal bovine serum), added to a magnetic pole and allowed to stand for 2 minutes, then carefully discarded the supernatant. Repeat the above process 4 times. Take the magnetic beads after washing, add the magnetic beads to PBMC cells, mix well, and culture them at 37 degrees for 3 days. After 3 days, the magnetic beads were taken out, and the T cells were first resuspended several times with a pipette gun. Place the cell suspension in the magnetic pole, and after standing for two minutes, discard the magnetic beads on the tube wall. Count again, and the counting results are shown in Table 2.
[0104] Table 2
[0105] T cell density 1.9×10 6 pcs / ml...
Embodiment 3
[0106] Example 3: Adeno-associated virus vector construction
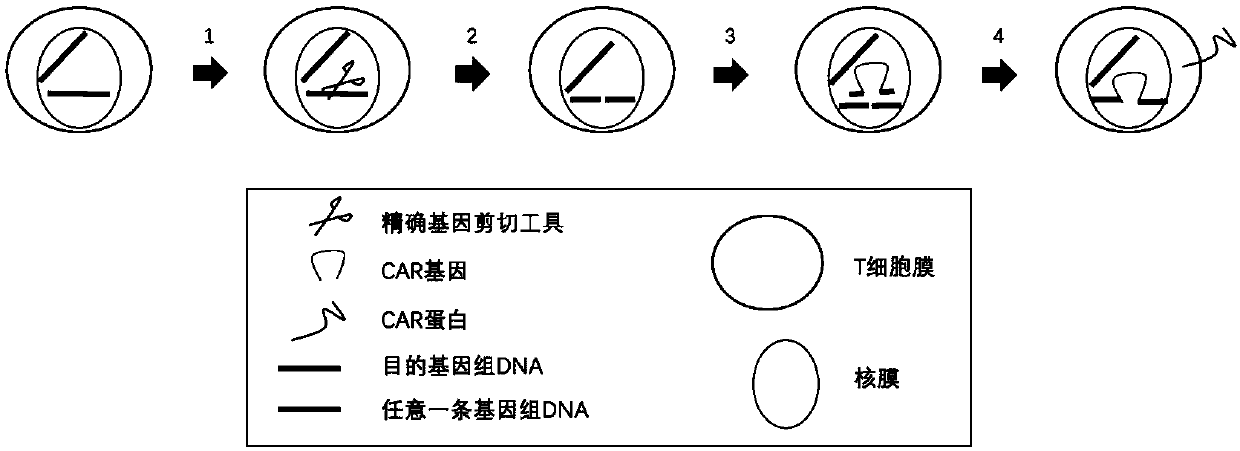
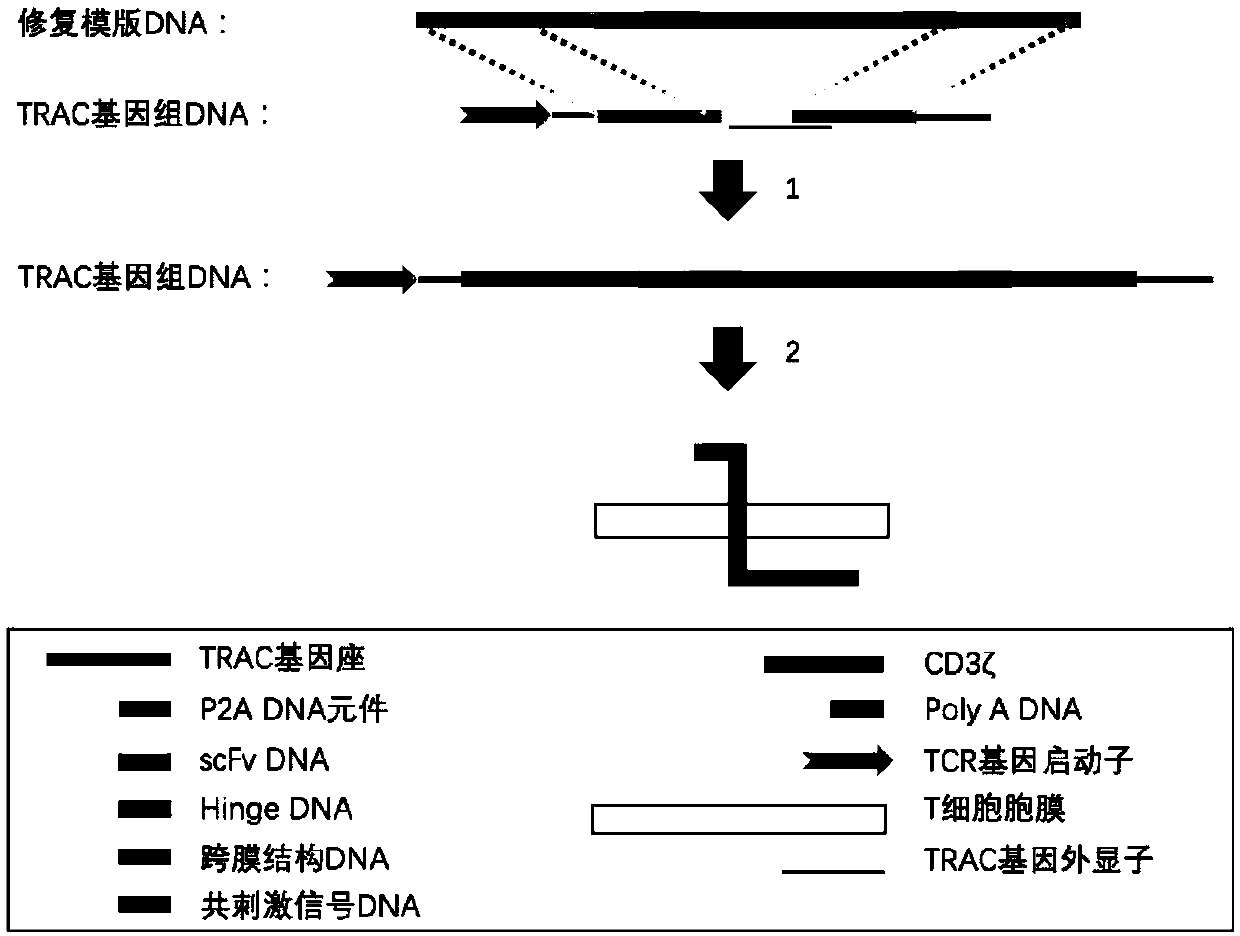
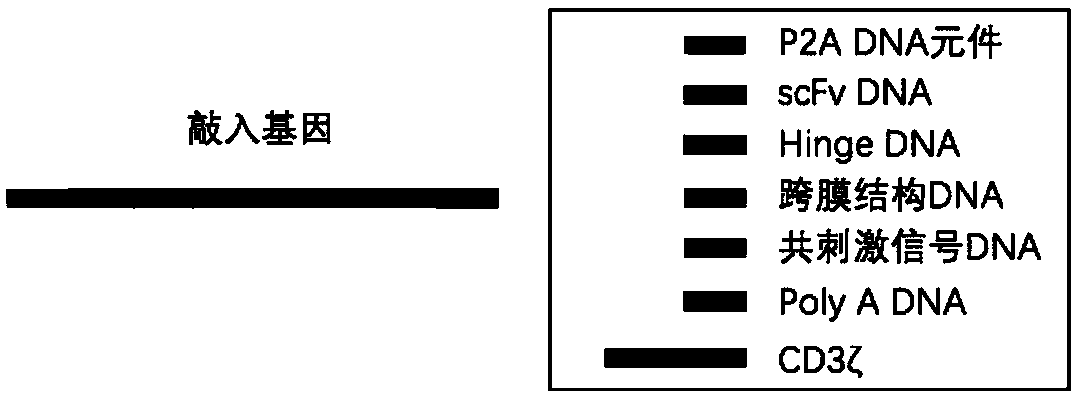
[0107] In the present invention, the CAR gene is knocked into the TRAC gene locus using the method of CRISPR-Cas9 technology combined with adeno-associated virus vector transduction. Firstly, CRISPR-Cas9 is used to make a cut at the TRAC gene site, and then the CAR gene template delivered by adeno-associated virus is used to repair the gap by homologous recombination, and finally the CAR gene is knocked into the target site. Schematic such as figure 2 .
[0108] (1) The adeno-associated virus vector comprises two parts:
[0109] a. Left and right homology arms. The left and right homology arms are used to identify the target DNA and carry out recombination and exchange. In order to ensure the correct expression of the knock-in gene, the present invention "logs in" the knock-in gene to the 5' end region of exon 1 of the TRAC gene. According to the length of the base sequence, the following pairs of homology arm...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com