Bacillussubtilis CRISPR-Cas9 genome editing system
A Bacillus subtilis genome editing technology, applied in the field of genome editing, can solve the problems of low accuracy, low editing efficiency, and unrealized iterative cycle system, and achieve high accuracy and realize the effect of iterative editing cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
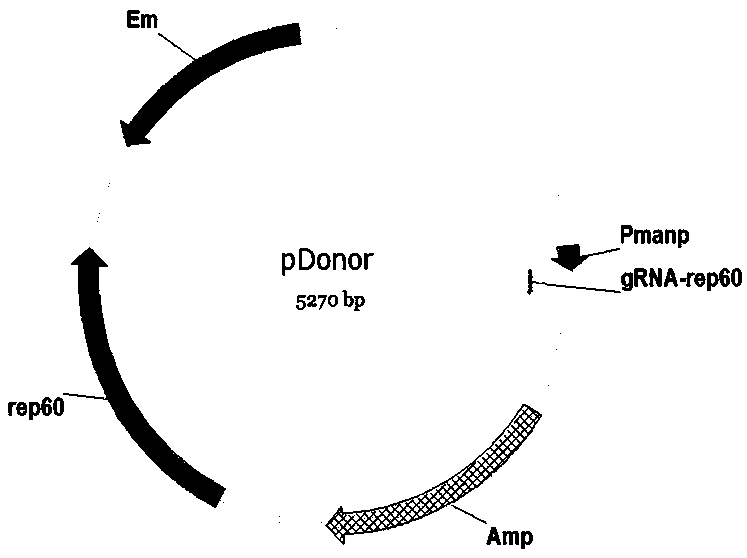
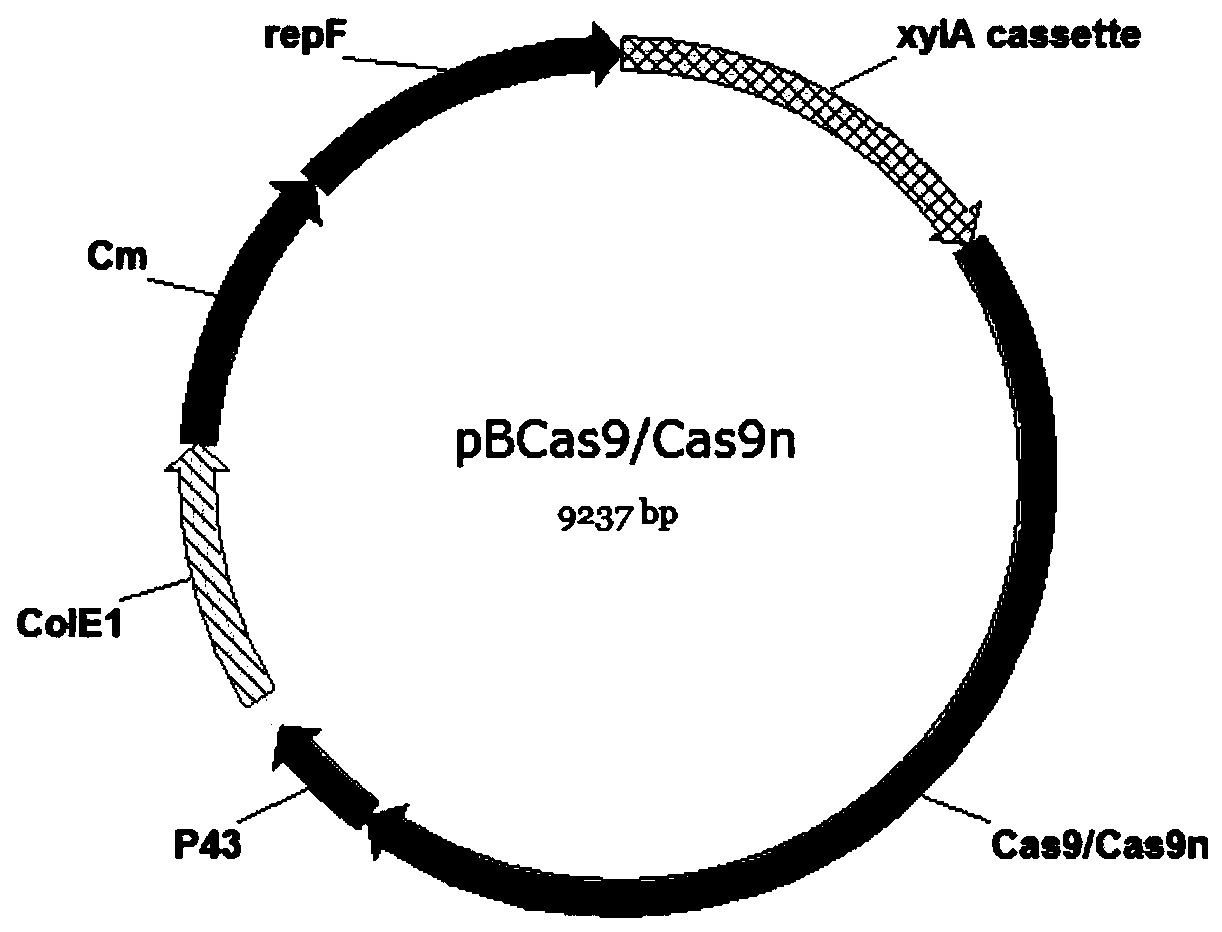
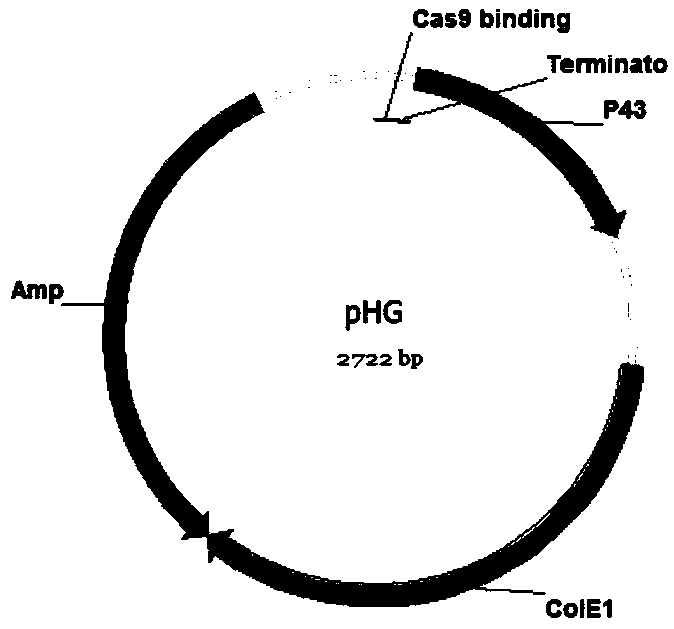
[0047] Example 1 Construction of tool plasmids pBSCas9, pBSCas9n, pDonor and pHG
[0048] The pBSCas9, pBSCas9n and pDonor tool plasmids are used in the operation process of the present invention, and the specific construction method is as follows.
[0049] (1) Construction of tool plasmid pBSCas9 and pBSCas9n
[0050] Using B. subtilis 168 genome as template, using primers P43-F and P43-L to amplify the P43 promoter sequence (SEQID NO.31), using plasmid pUC18 as template, using primers ColE1-F and ColE1-L to amplify pUC18 replication Sub-ColE1 sequence, using primers P43-F and ColE1-L to fusion the above two fragments into fragment F1. In this step of fusion, the sequence of gRNA binding to Cas9 protein and terminator sequence are introduced downstream of the P43 promoter through primers. Using plasmid pHP13 as a template, using primers Cm-F and Cm-L to amplify the chloramphenicol resistance gene and its promoter sequence (Cm), using plasmid pEBs-cop1 as template, using primers re...
Embodiment 2
[0065] Example 2: Principle of CRISPR-Cas9 system gene editing
[0066] The principle of CRISPR-Cas9 system gene editing is as follows Figure 4 Shown.
[0067] (1) Principle of unit point editing
[0068] After determining the genome modification site, select the appropriate PAM site in the region to be modified, and determine the gRNA sequence (the 20bp sequence upstream of the PAM site is the gRNA sequence). The gRNA sequence was inserted into the downstream region of the P43 promoter of the pBSCas9 plasmid. The specific method is to design primers to perform full-plasmid amplification PCR on plasmid pBSCas9, introduce BsaI restriction sites and a part of gRNA sequence into the upstream and downstream primers, and reconnect them into circular plasmids through the Golden Gate cloning reaction. In this process, insert gRNA into P43 Downstream of the promoter (SEQ ID NO. 31), this process does not introduce any extra bases.
[0069] According to the PAM site and the type of gene edi...
Embodiment 3
[0077] Example 3: Principles of CRISPR-Cas9n system genome editing
[0078] The principle of CRISPR-Cas9n system gene editing is as follows Figure 4 Shown.
[0079] (1) Principle of unit point editing
[0080] After determining the genome modification site, select the appropriate PAM site in the region to be modified, and determine the gRNA sequence (the 20bp sequence upstream of the PAM site is the gRNA sequence). The gRNA sequence was inserted into the downstream region of the P43 promoter of the pBSCas9n plasmid. The specific method is to design primers to perform full-plasmid amplification PCR on plasmid pBSCas9n, introduce BsaI restriction sites and a part of gRNA sequence into the upstream and downstream primers, and reconnect them into circular plasmids through the Golden Gate cloning reaction. In this process, insert gRNA into P43 Downstream of the promoter (SEQ ID NO. 31), this process does not introduce any extra bases.
[0081] According to the PAM site and the type of g...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com