Method for analyzing phylogenetic development of babylonia formosae based on complete mitochondrial genomes
A mitochondrial genome and phylogeny technology, applied in the fields of genomics, biochemical equipment and methods, proteomics, etc., can solve problems such as confusion, misclassification, and vulnerability to environmental factors, and achieve convenient and effective analysis.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
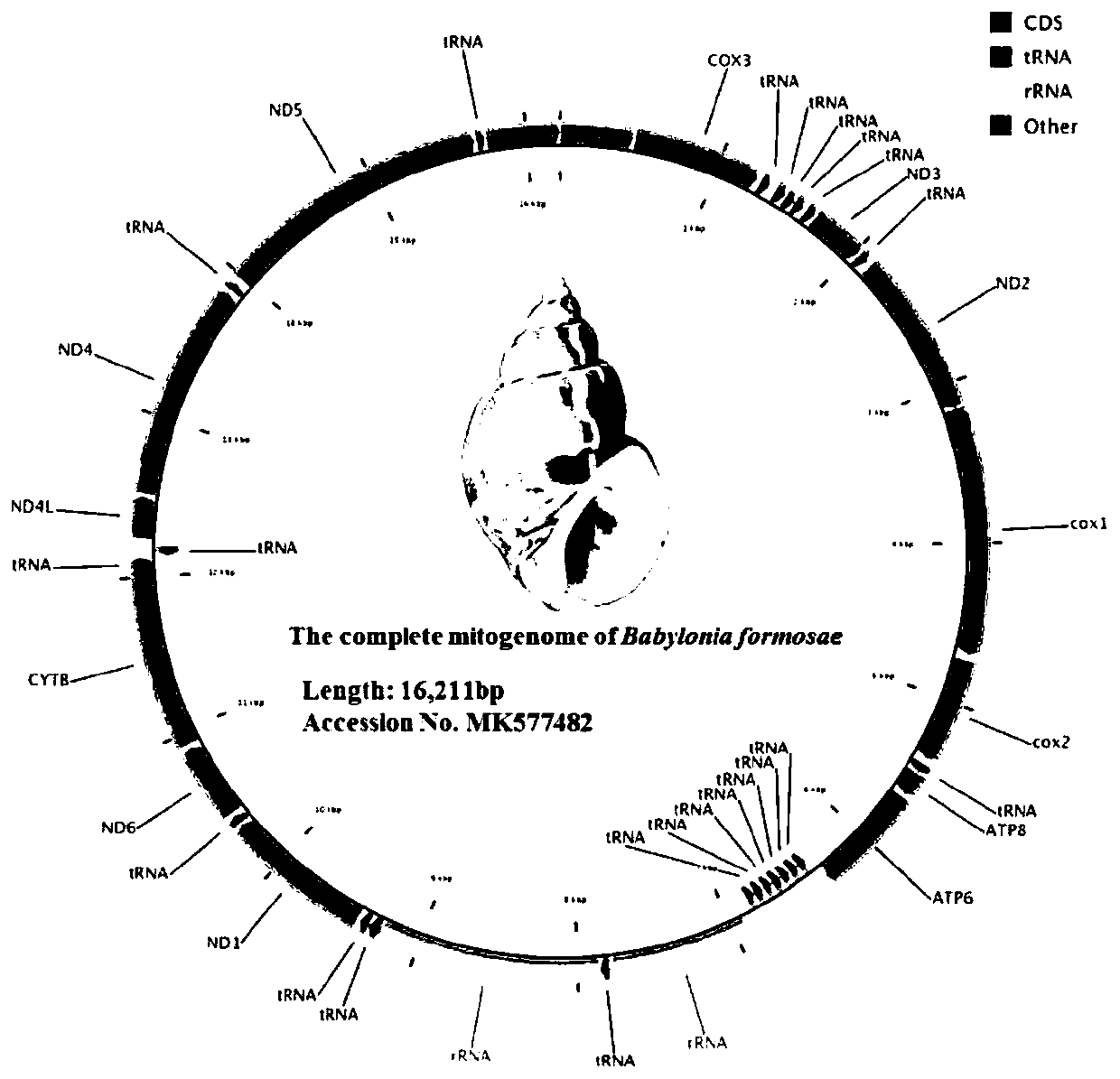
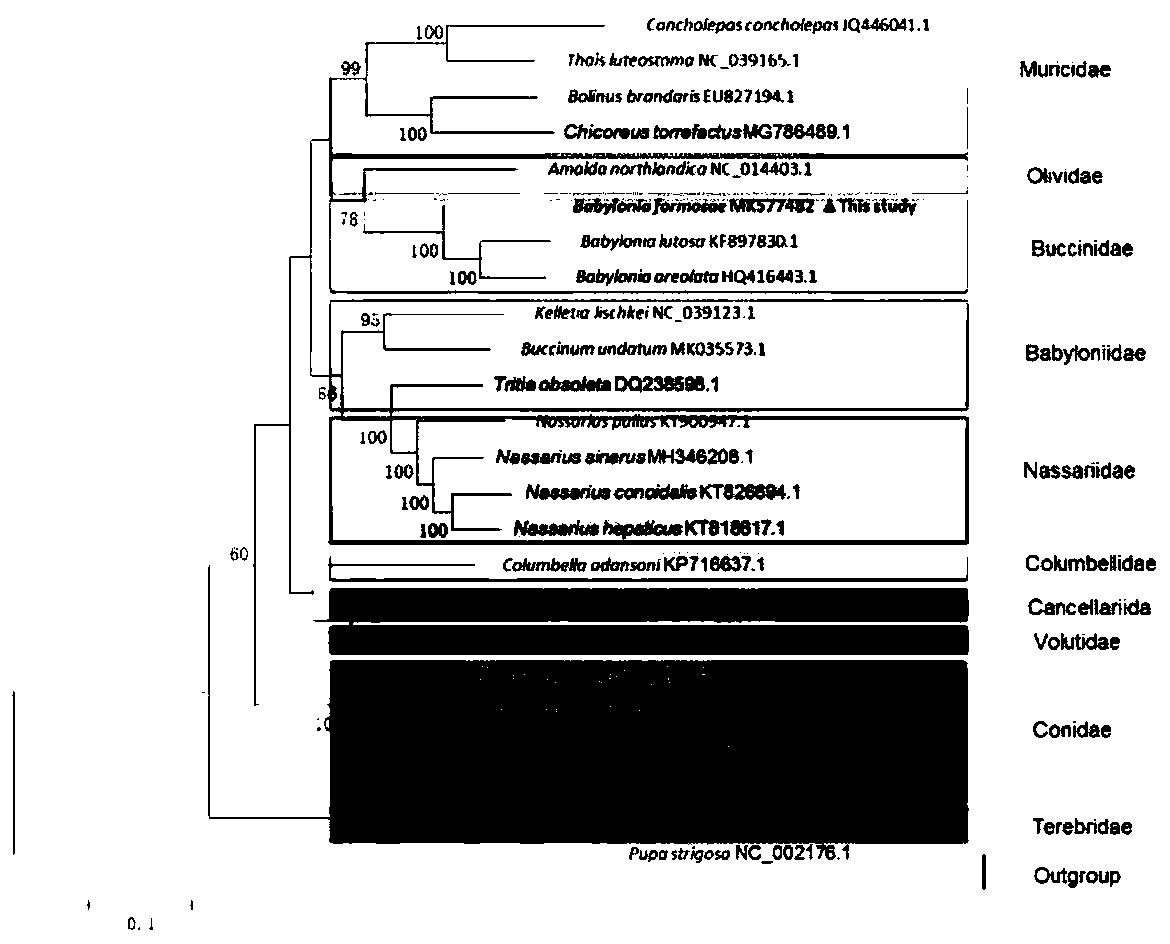
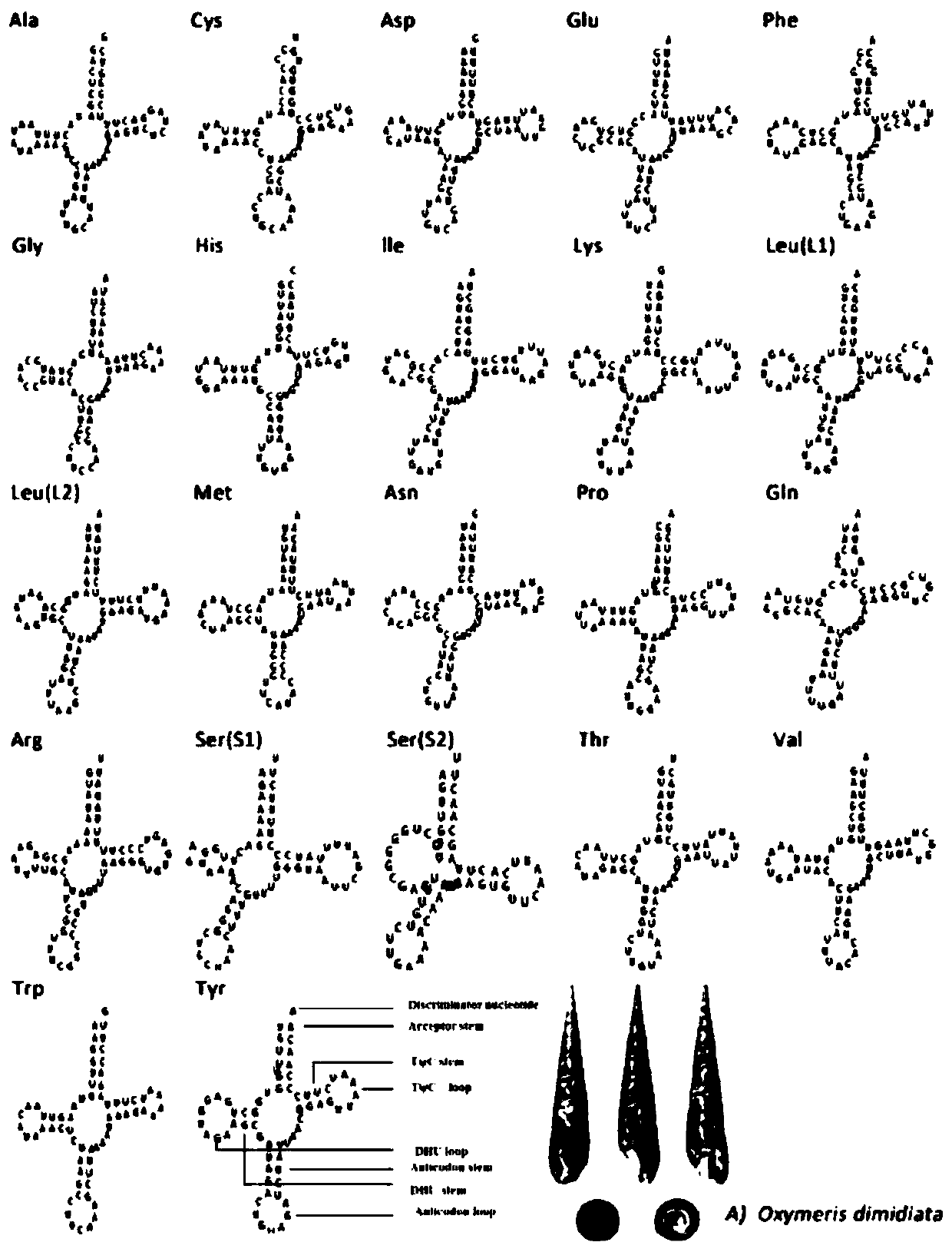
[0037] Embodiment 1: A method for analyzing the phylogeny of Taiwan Dongfeng snail based on the whole mitochondrial genome, comprising the following steps:
[0038] 1) Sample collection: Collect Taiwan Dongfeng snail, take 17mg Taiwan Dongfeng snail muscle tissue, add 450ul lysate, add 13ul Protein K, turn over, 57 ℃ water bath for 13h, then add 300ul NaCl solution with a concentration of 6M, vortex fully , placed at 4°C, centrifuged at 11,000 rcf, took the supernatant, added an equal volume of isopropanol, turned it over, and stored it in a freezer, then centrifuged and discarded the supernatant, dried the precipitate in an oven at 50°C after washing, and added 55ul of pure water Finally, the genomic DNA was detected by agarose gel electrophoresis; then the mitochondrial genomic DNA was extracted from the muscle tissue, frozen and stored at -25°C;
[0039] 2) Put the mitochondrial genomic DNA in an ultrasonic wave at 90W for 3s, repeat 4 times, and interrupt at an interval of...
Embodiment 2
[0044] Embodiment 2: A method for analyzing the phylogeny of Taiwan Dongfeng snail based on the whole mitochondrial genome, comprising the following steps:
[0045] 1) Sample collection: Collect Taiwan Dongfeng snail, take 15mg Taiwan Dongfeng snail muscle tissue, add 400ul lysate, add 10ul Protein K, turn over, 60 ℃ water bath for 12h, then add 300ul NaCl solution with a concentration of 6M, vortex fully , placed at 4°C, centrifuged at 10,000 rcf, took the supernatant, added an equal volume of isopropanol, turned it over, and stored it in a freezer, then centrifuged and discarded the supernatant, dried the precipitate in an oven at 45°C after washing, and added 50ul of pure water Finally, the genomic DNA was detected by agarose gel electrophoresis; then the mitochondrial genomic DNA was extracted from the muscle tissue and frozen at -30°C for cryopreservation;
[0046] 2) Put the mitochondrial genomic DNA in an ultrasonic wave under 80W for 2s, repeat 5 times, and interrupt a...
Embodiment 3
[0051] Embodiment 3: A method for analyzing the phylogeny of Taiwan Dongfeng snail based on the whole mitochondrial genome, comprising the following steps:
[0052] 1) Sample collection: Collect Taiwan Dongfeng snail, take 20mg Taiwan Dongfeng snail muscle tissue, add 500ul lysate, add 15ul Protein K, turn over, 55 ℃ water bath for 14h, then add 300ul NaCl solution with a concentration of 6M, vortex fully, Place at 4°C and centrifuge at 12000rcf, take the supernatant, add an equal volume of isopropanol, turn it over, and store it in a freezer, then centrifuge and discard the supernatant, wash the precipitate and dry it in an oven at 55°C, add 60ul of pure water , using agarose gel electrophoresis to detect genomic DNA; then extract mitochondrial genomic DNA from muscle tissue, freeze and store at -15°C;
[0053] 2) Put the mitochondrial genomic DNA in an ultrasonic wave under 100W for 4s, repeat 3 times, and interrupt at an interval of 10s, after purification, perform DNA end ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com