Granulocyte or monocyte marking system, and marking method and application thereof
A mononuclear cell and labeling system technology, applied in the field of granulocyte or monocyte labeling system, can solve the problem of inability to achieve specific expression of Cre recombinase
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Example 1 Determination of the Ms4a3 gene as a gene specifically expressed in granulocytes and monocyte precursors
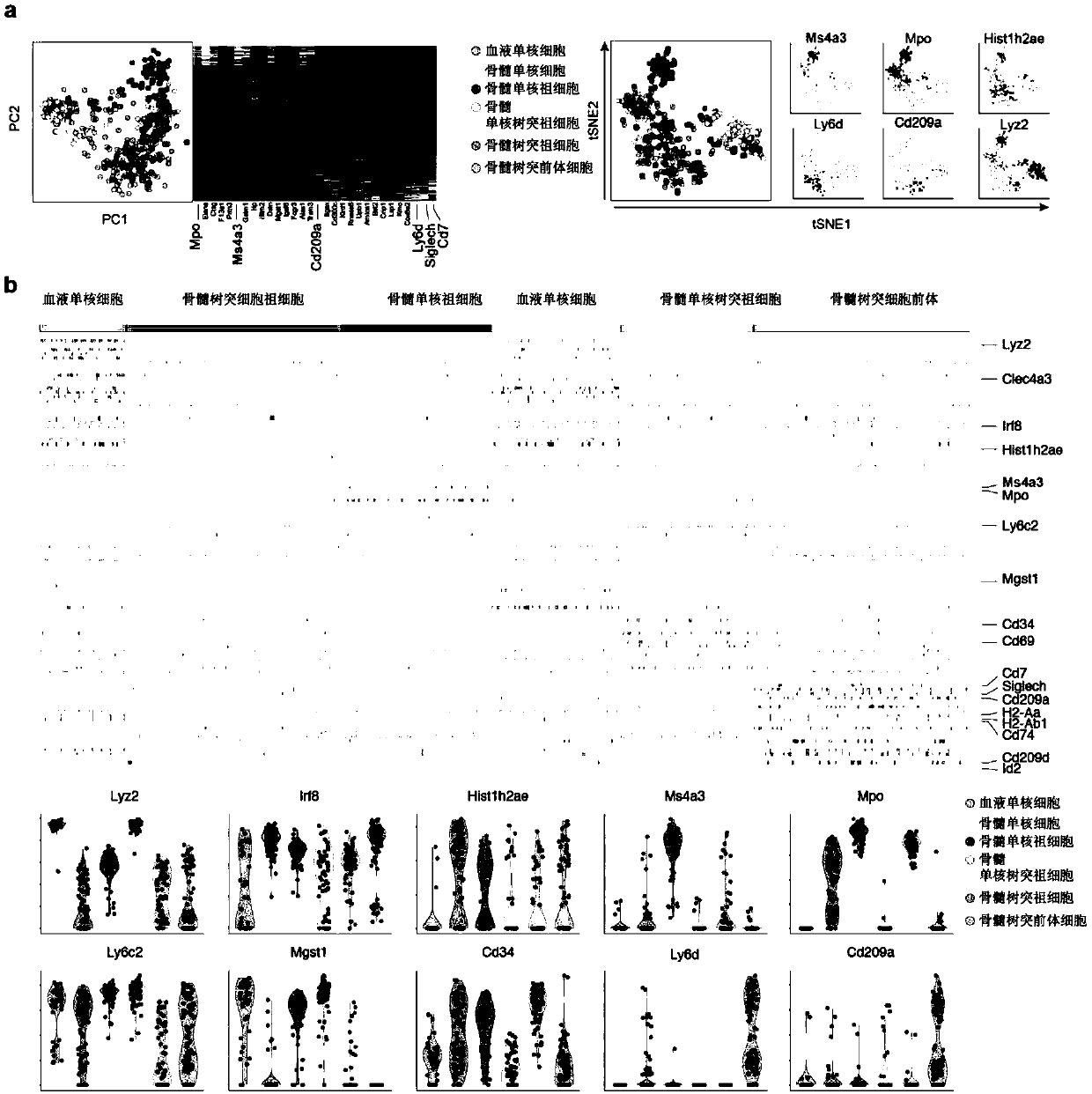
[0064] To identify genes specifically expressed in granulocyte and monocyte precursors, the present invention sorted blood monocytes / bone marrow mononuclear cells / bone marrow mononuclear cell precursors / bone marrow mononuclear-dendritic precursors / bone marrow Dendritic cell precursors, and use the C1Fluidigm single-cell platform to prepare a single-cell library for single-cell sequencing.
[0065] Experimental results such as figure 1 As shown, a, after the single-cell transcriptome data is analyzed by tSNE dimensionality reduction, the cells are clustered into different clusters (Cluster). Ms4a3 is specifically expressed in cMoP clusters. b, Heat map showing Ms4a3 expression in cMoP. c, Violin diagram shows the comparison of the expression of different genes in each cell population, among which Ms4a3 presents the most specific expression.
Embodiment 2
[0066] Example 2 Construction of the Ms4a3-Cre granulocyte-monocyte labeling system of the present invention
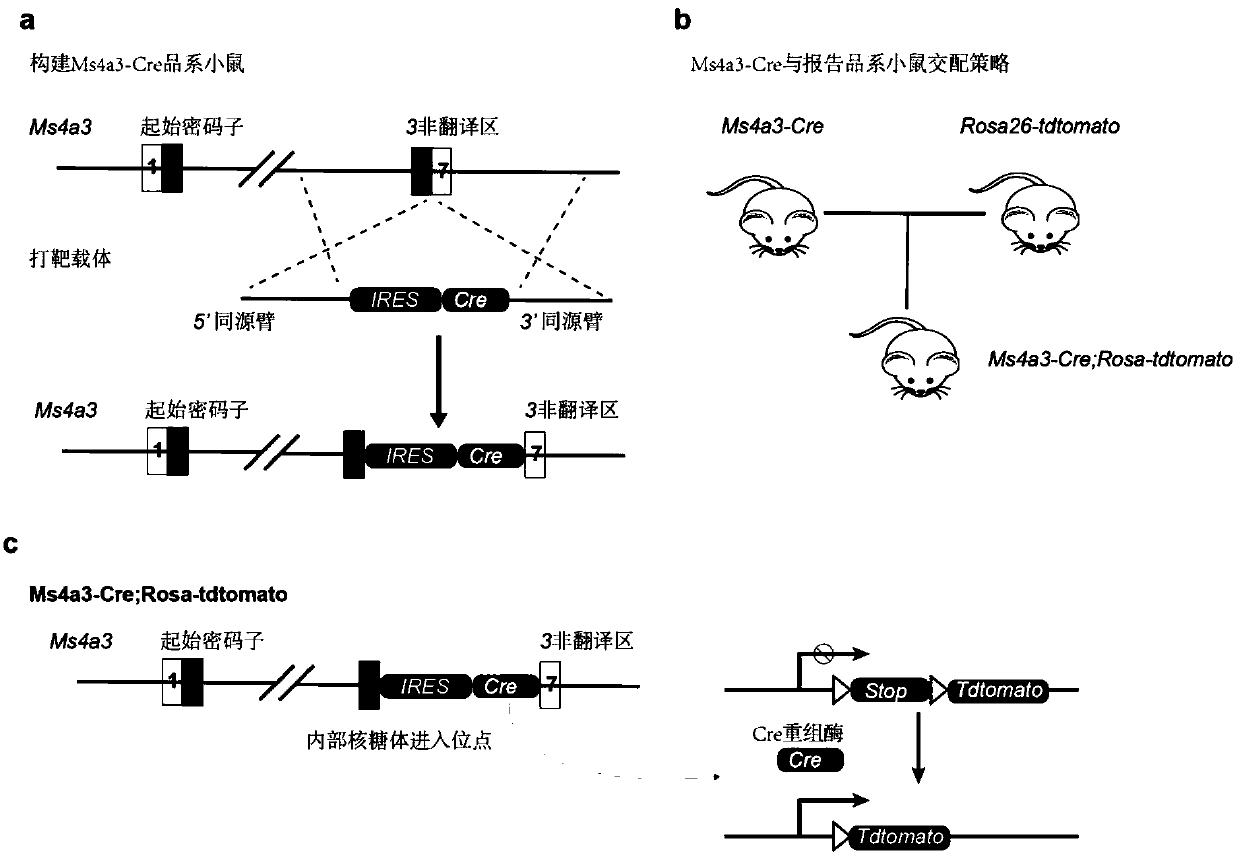
[0067] build method like figure 2 shown, including the following steps:
[0068] Step a, using the Crispr-Cas9 gene editing method to insert the IRES-Cre sequence after the stop codon of Ms4a3.
[0069] Step b, Ms4a3-Cre mice were crossed with Rosa26-tdTomato reporter strain mice to obtain Ms4a3-Cre; Rosa-tdTomato tracer mice.
[0070] In the present invention, the working principle of the tracer system is: in the granulocyte-monocyte precursor expressing Ms4a3, the Cre recombinase is expressed simultaneously with Ms4a3, and the Cre recombinase recognizes the Stop sequence at the Rosa26 site and removes it, so that tdTomato starts Therefore, the cell and its progeny cells will be labeled with tdTomato, with red fluorescence, which can be detected by flow cytometry and fluorescence microscopy.
Embodiment 3
[0071] Example 3 Ms4a3-Cre; Verification of Rosa-tdTomato granulocyte-monocyte labeling system
[0072] Specific steps are as follows
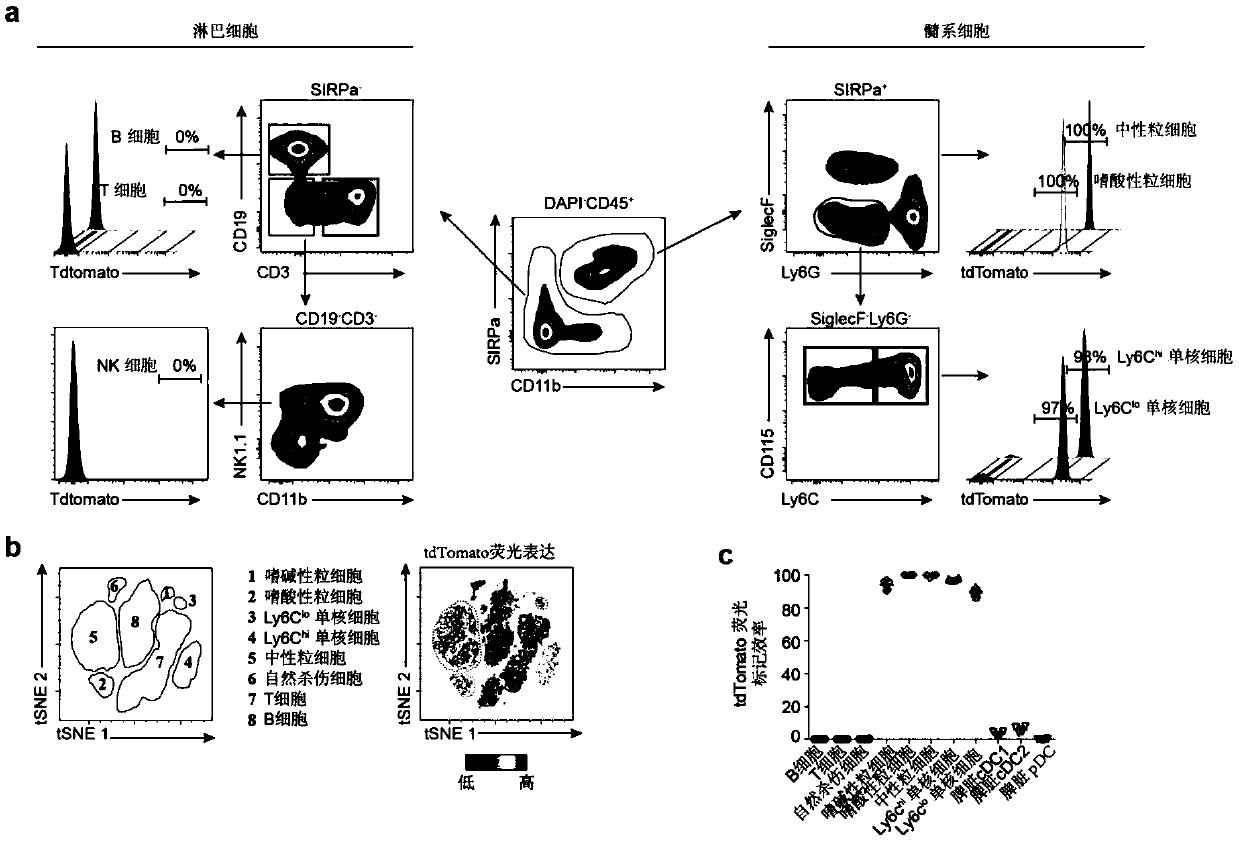
[0073] In step a, the peripheral blood of the mouse is taken, and the red blood cells are lysed to detect the marker status of each cell subgroup by flow cytometry.
[0074] In step b, the spleen of the mouse is taken, ground into a single cell suspension, and the red blood cells are lysed to detect the dendritic cell markers by flow cytometry.
[0075] Experimental results such as image 3 As shown, the Ms4a3 tracer model mouse blood cell lineages and the marker analysis of spleen dendritic cells. in, image 3 The a and b in the figure indicate the markers of the three lineages in the peripheral blood of the model mice detected by flow cytometry, and the granulocytes and monocytes were efficiently marked, and the lymphocytes such as T cells, B cells and NK cells were not marked; in the spleen The dendritic cells have only very low labelin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com