K-ras gene mutation site detection kit
A detection kit, 1.k-ras technology, applied in the fields of molecular biotechnology and gene detection, can solve the problems of high operator requirements, high cost of kits, and difficulty in mutating genes, and achieve low-cost detection and detection. High efficiency and avoid cross-contamination effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] The primer probe that embodiment 1 K-ras gene mutation site detection uses
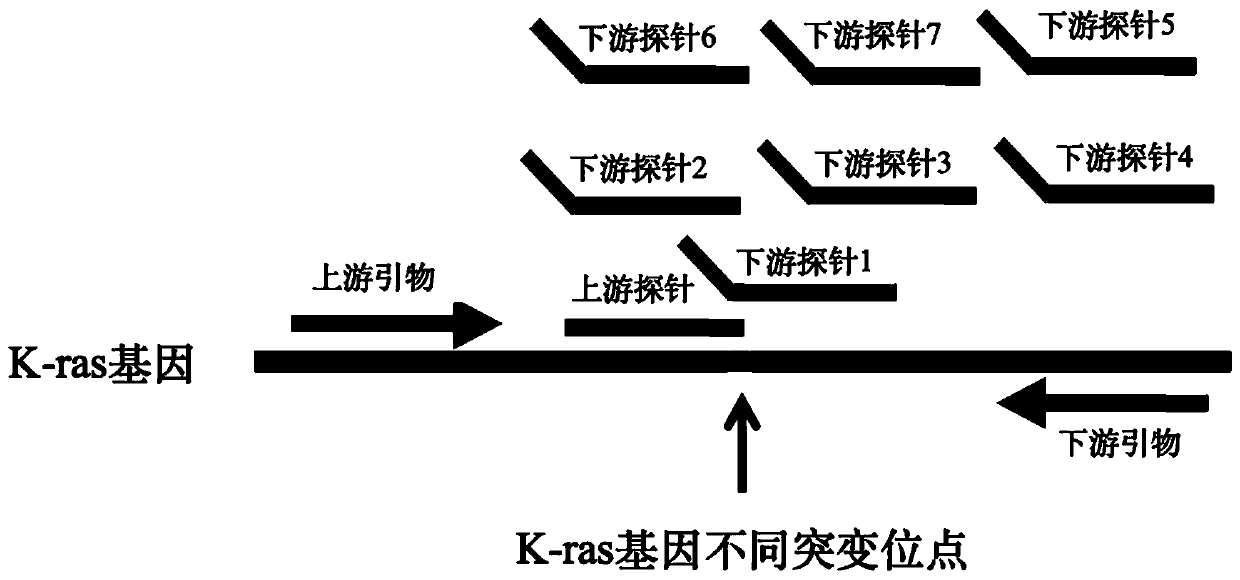
[0050] Design specific primers and probes according to the sequence of the hot spot mutation site (as shown in Table 1) on exon 12 and 13 of the K-ras gene, and the specificity of the detection results can be determined jointly by the primer pair and the probe pair. Design principles such as figure 1 shown.
[0051] Table 1. Common K-ras gene hotspot mutation types
[0052]
[0053] The K-ras gene mutation site detection kit consists of primers, conventional probes, nano gold probes, reaction solution and enzyme solution, and the sequences of the primers and probes are as follows.
[0054] Upstream primer: GTA CTG GTG GAG TAT TTG ATA GTG TAT TAA CCT TAT G (SEQ ID NO: 1);
[0055] Downstream primer: TGG TCC TGC ACC AGT AAT ATG CAT ATT AAA ACA AG (SEQ ID NO: 2);
[0056] Upstream probe: GGACACCGCATGGC (SEQ ID NO: 3);
[0057] Downstream probe 1: CACCGCATGGCTCACCAGCTCCAACTAC (SEQ ID NO: 4)...
Embodiment 2
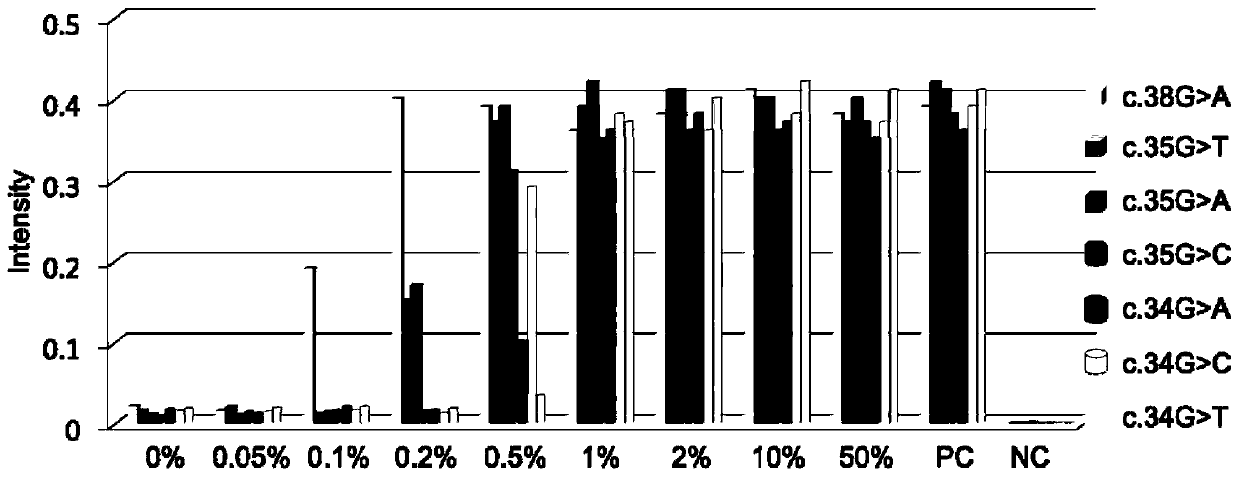
[0067] Example 2 Verification of independent detection results of seven mutation sites of K-ras gene
[0068] In this embodiment, the K-ras gene mutation site detection kit described above is used to detect Templates with different K-ras gene percentage mutation content are used to verify the feasibility of a single independent detection system.
[0069] The reaction volume of the kit is 20 μL, and its components are: 10 mM Tris buffer (pH 8.5), 1 μM primers (upstream primers, downstream primers), 1 μM probes (upstream probes, downstream probes 1-7, hair clip probe), 0.1 μM nano-gold probe (nano-gold probe 1, nano-gold probe 2), 0.2 mM dNTP, 0.5 U Taq DNA polymerase and endonuclease AfuFEN. The specific mutation templates to be tested are respectively added to the corresponding detection systems in the mixing ratios of 50%, 10%, 2%, 1%, 0.5%, 0.2%, 0.1%, 0.05%, and 0%. Perform negative (NC) and positive (PC) quality control at the same time, and the reaction program is: 95°C...
Embodiment 3
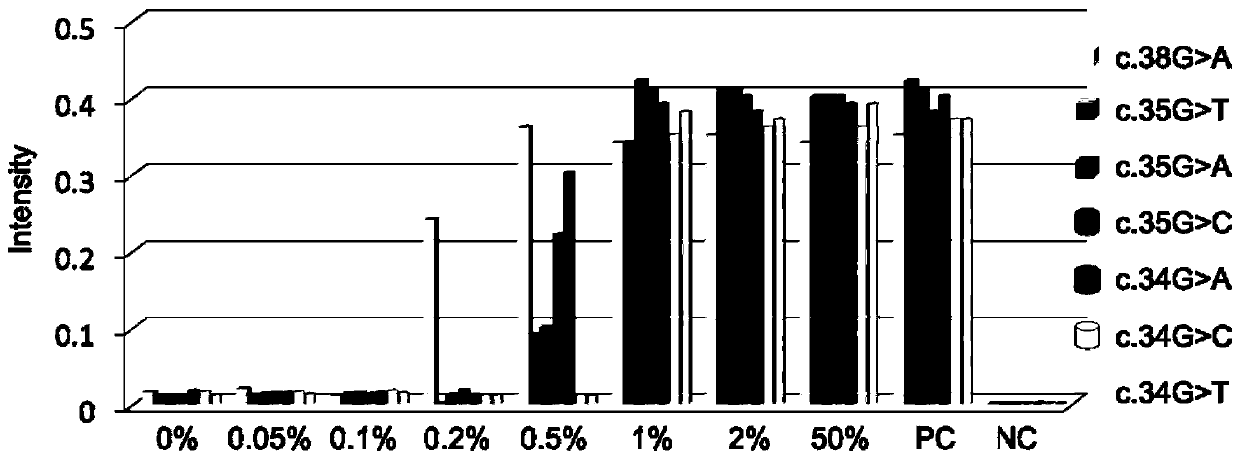
[0070] Example 3 Verification of the same tube detection results of seven mutation sites of K-ras gene
[0071] This embodiment uses the K-ras gene mutation site detection kit described above, using a pair of primers, probe 1, nano-gold probe, reaction solution, enzyme solution, probe 2-probe 8, etc. to detect different The template of K-ras gene percentage mutation content is used to verify the feasibility of multiplex detection for detecting multiple mutation sites in a single reaction system.
[0072] The reaction volume of the kit is 20 μL, and its components are: 10 mM Tris buffer (pH 8.5), 1 μM primers (upstream primers, downstream primers), 1 μM probes (upstream probes, downstream probes 1-7, hair clip probe), 0.1 μM nano-gold probe (nano-gold probe 1, nano-gold probe 2), 0.2 mM dNTP, 0.5 U Taq DNA polymerase and endonuclease AfuFEN. The specific mutation templates to be tested are respectively added to the corresponding detection systems in the mixing proportions of 5...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle size | aaaaa | aaaaa |
| particle size | aaaaa | aaaaa |
| particle size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com