Method and device for identifying microbial contamination in second-generation targeted capture sequencing sample
A technology of microbial contamination and targeted capture, applied in the fields of gene sequencing and bioinformatics, can solve problems such as microbial contamination, affecting the quality of human clinical nucleic acid sequencing libraries, and failure of human clinical sample sequencing, so as to reduce experimental workload and improve quality Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
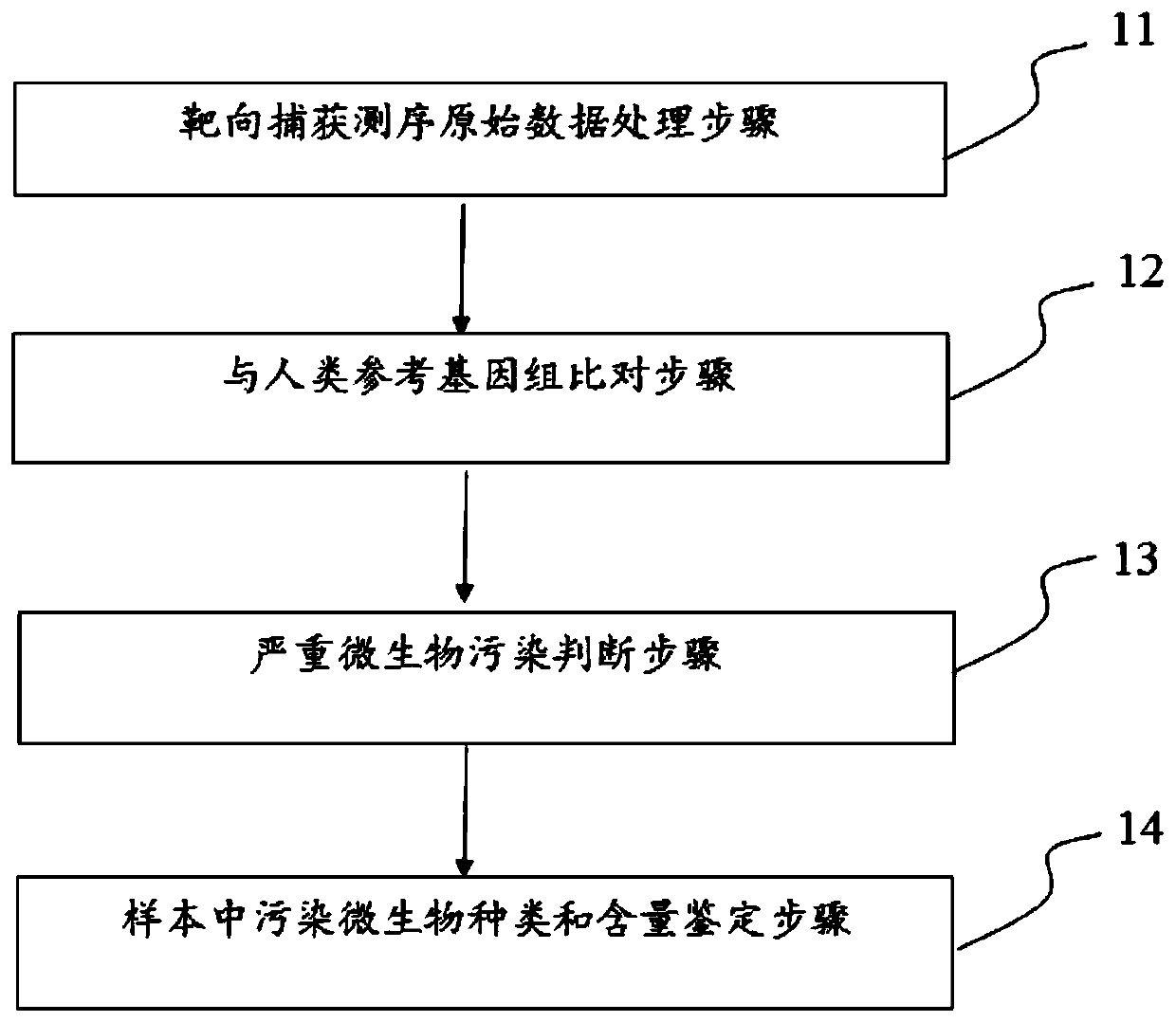
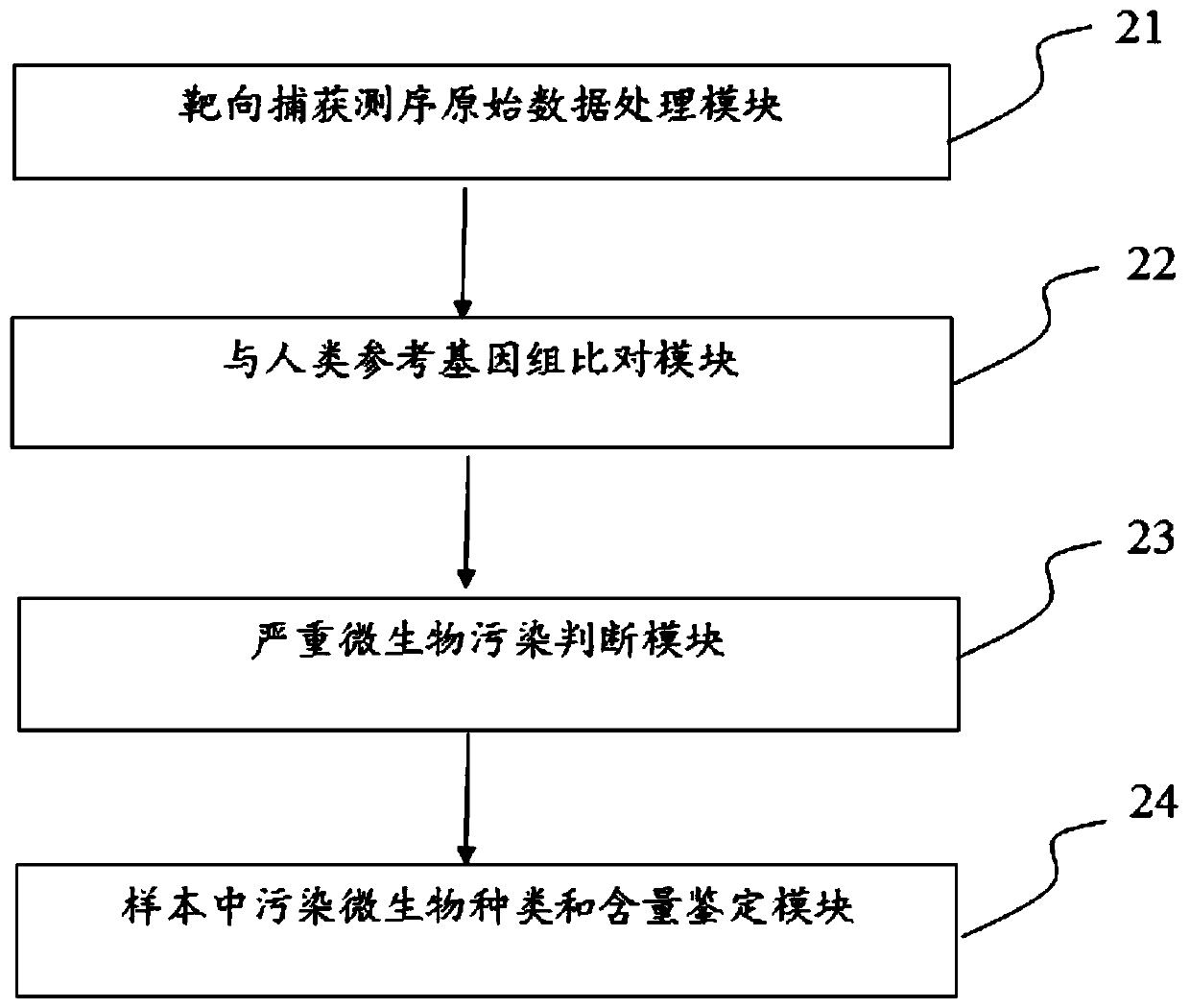
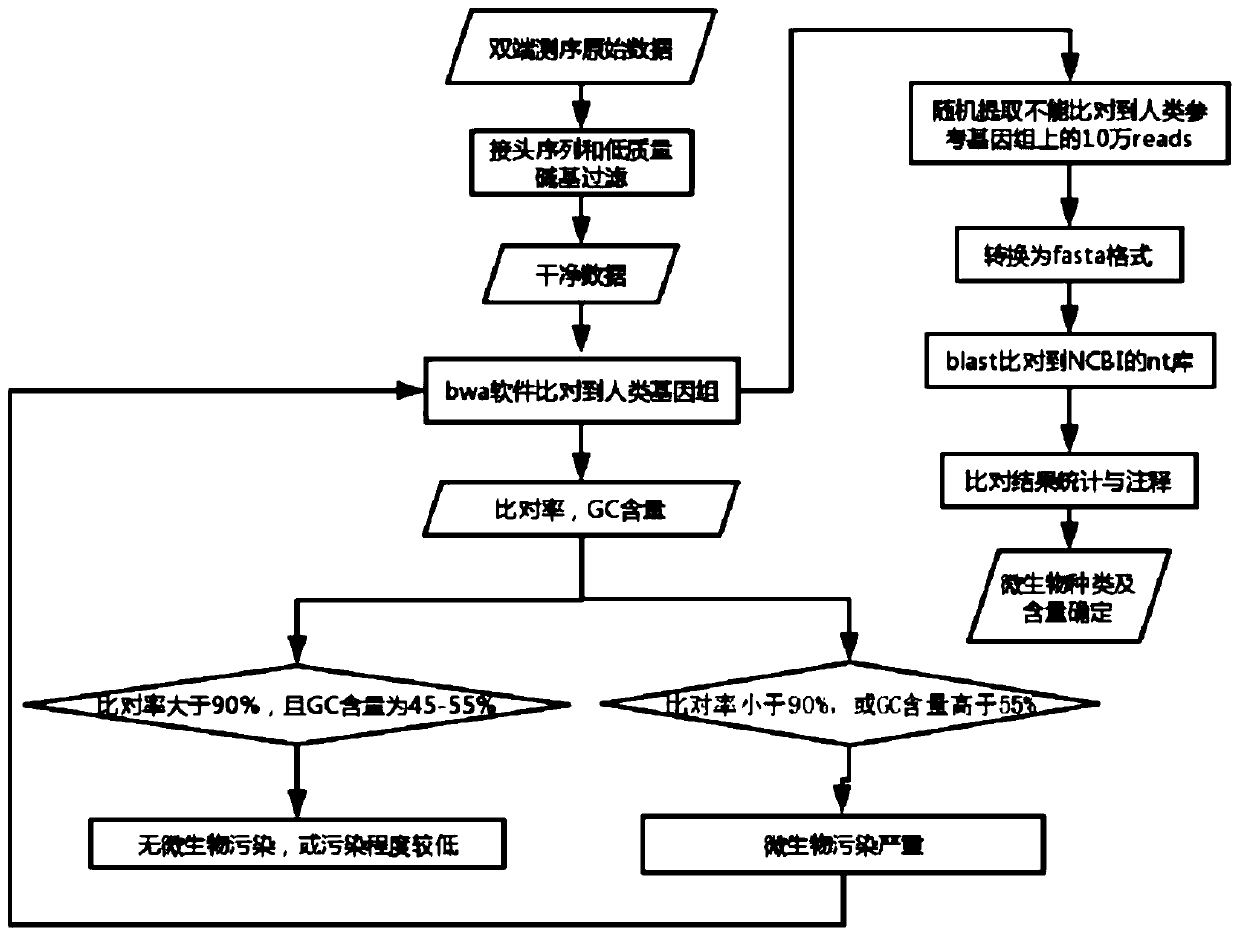
[0048] A 10ml venous peripheral blood sample was obtained from a patient with advanced lung cancer, and the second-generation library was constructed. DNA sequencing was performed on the NovaSeq6000 to obtain the original data of double-end sequencing, and the output was the original sequencing file fastq. Use the fastp software (https: / / github.com / OpenGene / fastp) independently developed by our company to perform adapter sequence removal, low-quality base filtering and data quality control on the original sequencing file fastq to obtain a clean fastq file.
[0049] Next, use the bwa software (https: / / github.com / lh3 / bwa) to compare the clean fastq file to the human reference genome hg19 (GRch37) to obtain the bam file, and further use the samtools software (https: / / github.com / samtools / ) for quality control analysis to obtain the comparison rate and GC content information of the sequencing data.
[0050] Then, the preset thresholds in the step of judging severe microbial contam...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com