Primer used for detecting nucleotide sequence variation, composition thereof, and method
A technology of nucleic acid sequence and target sequence binding region, applied in the field of molecular biology, to achieve the effect of solving cross-reaction problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
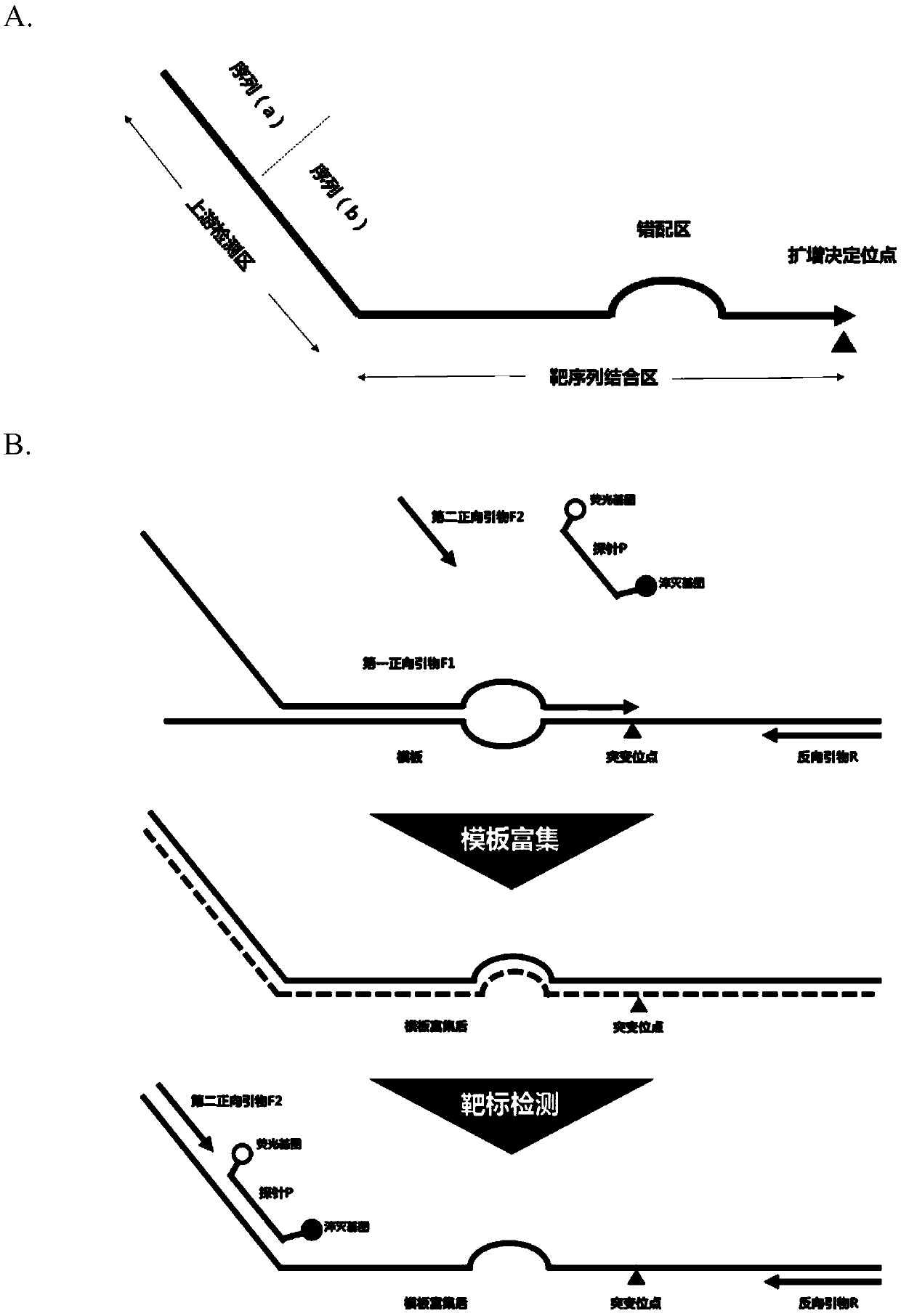
[0162] In this embodiment, a common nucleic acid variation is taken as an example to test the primers and detection system of the present invention. Specifically, the V600E mutation of the human BRAF gene was used as an example to simulate clinical samples to evaluate the performance of the detection system of the present invention.
[0163] BRAF mutations are found in about 8% of human tumors, and BRAF mutations can drive tumor proliferation, growth and differentiation, especially in colorectal cancer (5%-8%), thyroid cancer (5%-20%) And melanoma (40% ~ 68%), lung cancer, liver cancer and pancreatic cancer are also found to have different proportions of BRAF mutations. About 89% of the mutations occurred in the activation region of the 15th exon, most of which were the mutation of base T to A in the 1799th amino acid, resulting in the replacement of valine in BRAF by glutamic acid (V600E).
[0164] The detection of such point mutations is generally performed on the circulati...
Embodiment 2
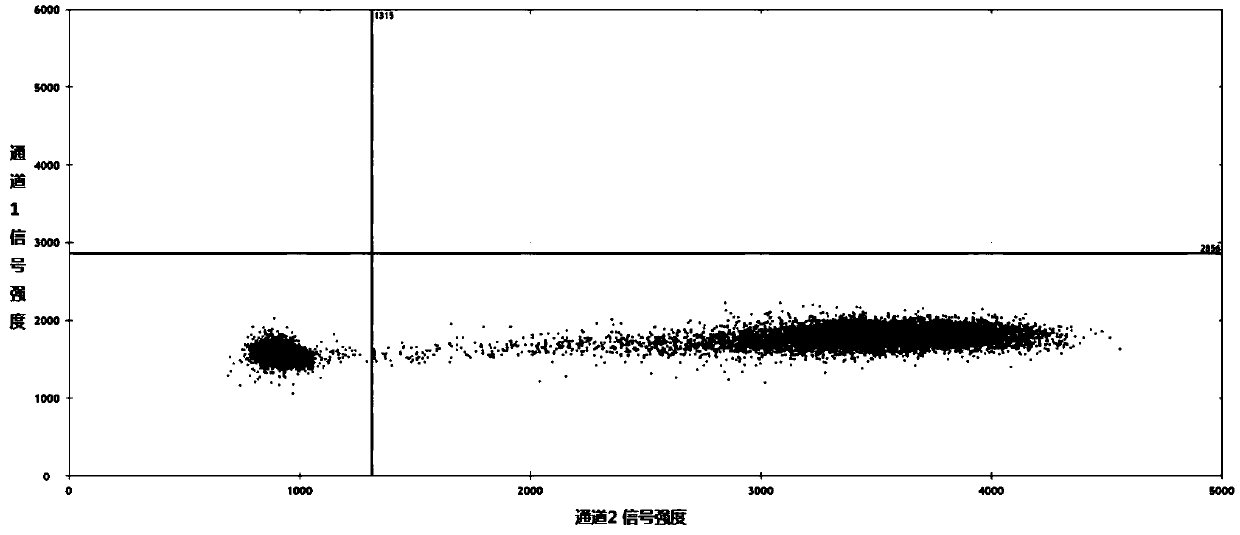
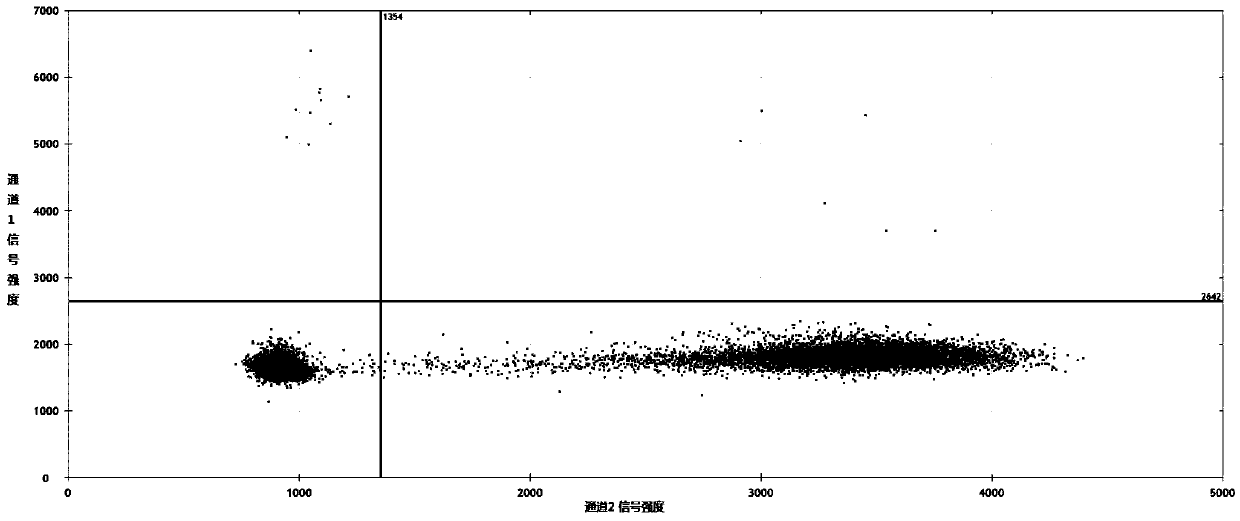
[0205] In this example, the test effects of kits from similar manufacturers were compared using DNA samples from the Colo 205 cell line (BRAF V600E mutation).
[0206] 1. Sample preparation: the same as in Example 1, except that a fragmented Colo 205 cell line DNA sample containing a BRAF gene V600E mutation with a mutation abundance of 65.8% was used, without serial dilution and preparation of the sample.
[0207] 2. Reaction system
[0208] The preparation of the primer system of the present invention is the same as in Example 1.
[0209] The comparison kit used "PrimePCR" from Bio-Rad TM ddPCR Mutation Assay Kit: BRAF WT for p.V600E, and BRAF p.V600E" commercially available kit (Catalog No. 1863100) was used as the comparison kit of the present invention, and the usage procedure was carried out according to its instruction manual, and the reaction preparation was shown in Table 3.
[0210] table 3
[0211]
[0212]
[0213] 3. Reaction Cell Preparation
[0214] Ad...
Embodiment 3
[0226] In this example, the detection effect of the F1 primer with a longer mismatch region was tested. The specific steps are as follows:
[0227] 1. Sample preparation: same as Example 1
[0228] 2. Preparation of reaction system
[0229] 2.1 Primers and probes
[0230] Primers and probes were synthesized by Sangon Bioengineering Co., Ltd.
[0231] Wherein, the specific sequences of the designed primers are as follows:
[0232] Table 4
[0233]
[0234] Wherein, the primer probes used are: mutant F1 (SEQ ID NO:8), wild type F1 (SEQ ID NO:9), mutant F2 (SEQ ID NO:3), wild type F2 (SEQ ID NO :4), mutant probe P (SEQ ID NO:5), wild type probe (SEQ ID NO:6) and reverse primer R (SEQ ID NO:7). The total length of the mutant F1 primer is 78bp, and the total length of the wild-type F1 primer is 77bp. The 39 bases at the 3' end of the two are the target nucleic acid binding region, the 3' end is the BRAF gene V600E mutation site, and the eighth base at the 3' end 14 mism...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com