Chlamydomonas reinhardtii instability domain gene, protein expression regulation method based on instability domain and application of chlamydomonas reinhardtii instability domain gene
A rhine coat and gene technology, applied in the field of genetic engineering, can solve the problems of low expression, silence, limited means, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0098] Example 1 Design and synthesis of Chlamydomonas reinhardtii instability domain gene crDi
[0099] Take the mutant rapamycin-binding protein containing point mutation sites F36V and L106P as the optimization object, select the codon with the highest frequency according to the codon preference of Chlamydomonas reinhardtii, and redesign the coding sequence; use ChlamySequence Optimizer (https: / / www.mathworks.com / matlabcentral / fileexchange / 65416-iddoweiner-coding-sequence-optimization-for-chlamydomonas-reinhardtii) software to further optimize the coding sequence, optimize the secondary structure of 5'UTR, and remove redundant potential content Containing the splicing site, obtaining the mutant type rapamycin binding protein gene shown in SEQ ID NO:2;
[0100] Calculated from the translation initiation site of the gene shown in SEQ ID NO: 2, the Chlamydomonas reinhardtii RBCS2 intron 1 (SEQ ID NO: 3 ), obtain the final optimized Chlamydomonas reinhardtii instability domai...
Embodiment 2
[0102] Example 2 Regulating the Expression of Chlamydomonas reinhardtii Aminoglycoside Phosphotransferase Using the Destabilizing Domain
[0103] (1) Construction of destabilizing domain-aminoglycoside phosphotransferase fusion gene recombinant vector
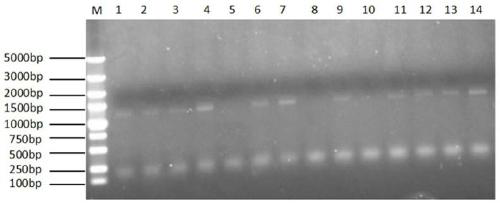
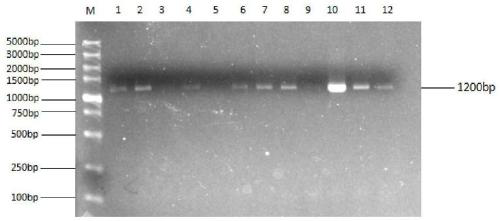
[0104] Using the synthesized crDi gene as a template, primers DParF (SEQ ID NO: 5) and DParR (SEQ ID NO: 6) were amplified at 62°C to obtain the destabilizing domain gene; the plasmid pOPt-mVenus-paro (source Based on the literature Targeted expression of nuclear transgenes in Chlamydomonas reinhardtii with aversatile, modular vector toolkit) as a template, primers ParF (SEQ ID NO: 7) and ParR (SEQ ID NO: 8) were used to amplify at 62°C to obtain aminoglycoside phosphotransfer Enzyme gene: using a mixture of 50ng of destabilizing domain gene and aminoglycoside phosphotransferase gene as a template, using primers DParF and ParR to amplify at 63°C to obtain the fusion gene crDi-pa;
[0105] After digesting the fusion gene crDi-p...
Embodiment 3
[0131] Example 3 Regulating the expression of Chlamydomonas reinhardtii luciferase by destabilizing domain
[0132] (1) Design and synthesis of Chlamydomonas reinhardtii luciferase gene
[0133] In this embodiment, the luciferase gene is first optimized, and Gaussia princeps luciferase (amino acid sequence shown in SEQ ID NO: 9) is used as the optimization object, and the codon with the highest frequency of use is selected according to the codon preference of Chlamydomonas reinhardtii ; Use Chlamy Sequence Optimizer (https: / / www.mathworks.com / matlabcentral / fileexchange / 65416-iddoweiner-coding-sequence-optimization-for-chlamydomonas-reinhardtii) software to further optimize the coding sequence and optimize the secondary of 5'UTR structure, remove redundant potential intron splicing sites; insert Chlamydomonas reinhardtii RBCS2 intron 1 (SEQ ID NO: 10 ), insert Chlamydomonas reinhardtii RBCS2 intron 2 (SEQ ID NO: 11), which has the effect of enhancing the expression of exogenou...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com