Application of chlamydomonas reinhardtii LRR (Leucine Rich Repeat) gene in regulating and controlling cadmium resistance of chlamydomonas reinhardtii
A reinhardtii and tolerance technology is applied in the application field of the Chlamydomonas reinhardtii LRR gene in regulating the cadmium tolerance of Chlamydomonas reinhardtii, which can solve the problems of unrelated research reports on the regulation of heavy metal cadmium metabolism, and achieve stable traits, The method is simple and the method is feasible
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1: Obtaining Chlamydomonas transformants with randomly inserted aphⅧ fragments by electroporation
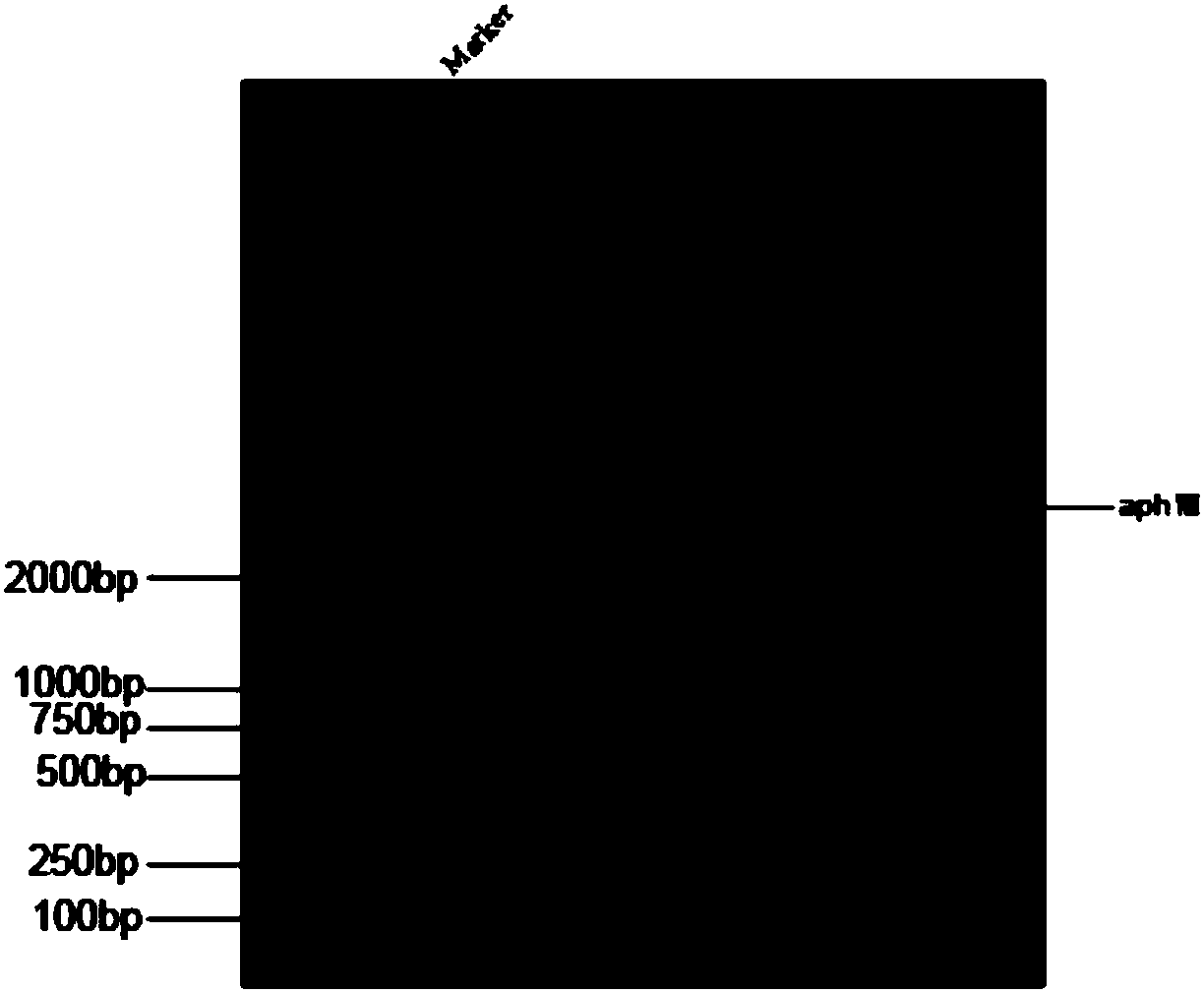
[0036] (1) Using the previously constructed plasmid pJMG-aphⅧ (Zhangfeng Hu, Yinwen Liang, Wei He, Junmin Pan * , Cilia Disassembly with Two Distinct Phases of Regulation, CellReports, 2015, 10(11): 1803-1810), the plasmid contains the aphⅧ fragment encoding paromomycin resistance, and the pJMG-aphⅧ plasmid is digested overnight ( EcoRI digestion), and then the digested plasmid was subjected to DNA agarose gel electrophoresis to separate the aphⅧ fragment (such as figure 1 shown), the gel was recovered to obtain the aphⅧ fragment (sequence shown in SEQ ID NO: 3).
[0037] (2) The wild-type Chlamydomonas 21gr was transferred from the TAP solid medium to the air-blown bottle containing the TAP liquid medium, and cultured for 3 days under continuous light conditions to make the concentration reach 1.5×10 7 cells / mL. The cultured cells were transferred to the Erlen...
Embodiment 2
[0042] Example 2: Screening transformants to obtain cadmium chloride-resistant mutant algal strains
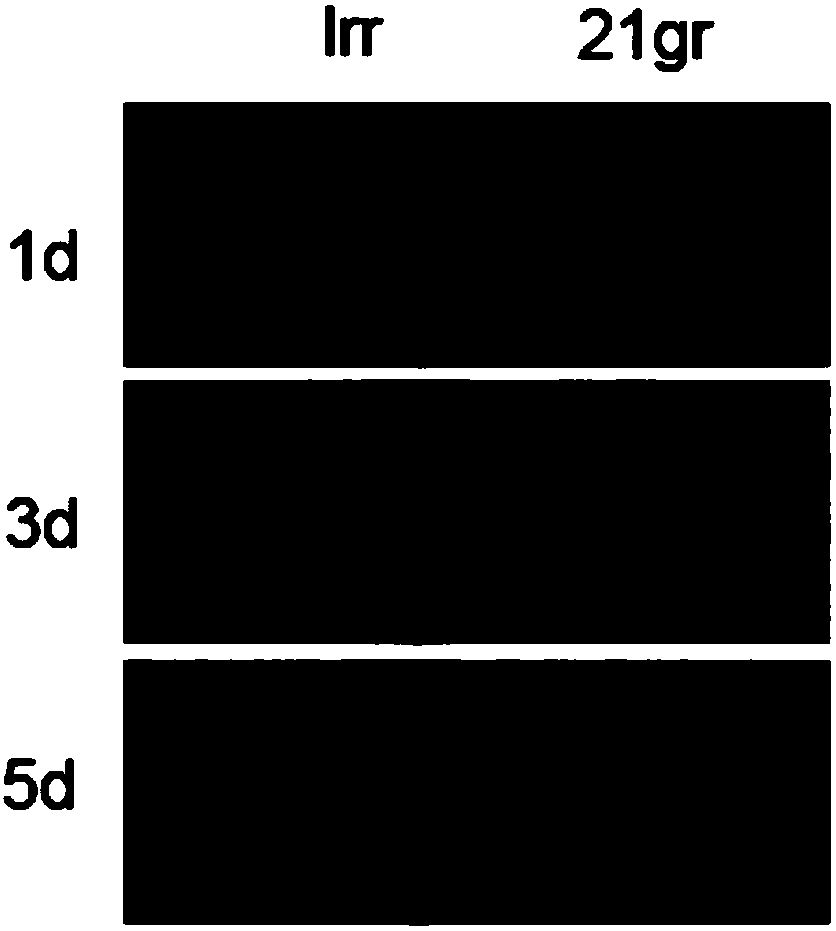
[0043] (1) Pick the Chlamydomonas transformants from the TAP solid medium containing paromomycin to the TAgP solid medium with a sterilized toothpick, and each transformant corresponds to a number. Then put it under the photoperiod and cultivate it for 3 days, and then pick it on the TAgP solid medium containing 0.6mM cadmium chloride, the numbers correspond to the numbers on the TAgP solid medium, and the wild type 21gr is used as a control. The screening concentration of 0.6mM cadmium chloride is the optimal screening concentration of heavy metal cadmium-resistant mutants obtained in the applicant's previous experiments. For details, see patent 201611022254.9.
[0044] (2) Put the 0.6mM cadmium chloride solid medium containing the transformants to cultivate under the photoperiod for 3-4 days, then observe and record the growth situation of the transformants on the 0.6mM cadm...
Embodiment 3
[0045] Example 3: Extracting the genome of the mutant
[0046] (1) Pick the mutant from the TAgP solid medium with a sterilized toothpick and culture it in the TAP liquid medium for 4 days.
[0047] (2) Collect the cells, centrifuge at 2500rpm for 3min, discard the supernatant, resuspend the cells with 4mL of liquid medium, transfer 1mL of the cells to a 1.5mL EP tube with a pipette gun, centrifuge at 14000rpm for 1min, discard the supernatant, and place the cells in Freeze in liquid nitrogen and store at -80°C for later use.
[0048] (3) Take out the frozen cells and add 650 μL of preheated CTAB lysate, pipette and mix the cells thoroughly, place them in a water bath at 65°C for 1 hour, and invert the EP tube every 10 minutes to fully lyse them.
[0049] (4) Add 650 μL of PCI (phenol: chloroform: isoamyl alcohol = 25:24:1). PCI should be shaken well before adding. After adding, invert the EP tube up and down to mix well, and centrifuge at 14,000 rpm for 10 min.
[0050] (5)...
PUM
| Property | Measurement | Unit |
|---|---|---|
| density | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com