Fluorescent chemical sensor for synchronously detecting multiple DNA (deoxyribonucleic acid) glycosylases and detection method and application of fluorescent chemical sensor
A technology of chemical sensor and glycosylase, which is applied in the field of fluorescent chemical sensor and its detection, can solve the problems of no specificity, harmful radiation, low sensitivity, etc., and achieve the effect of increased sensitivity and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
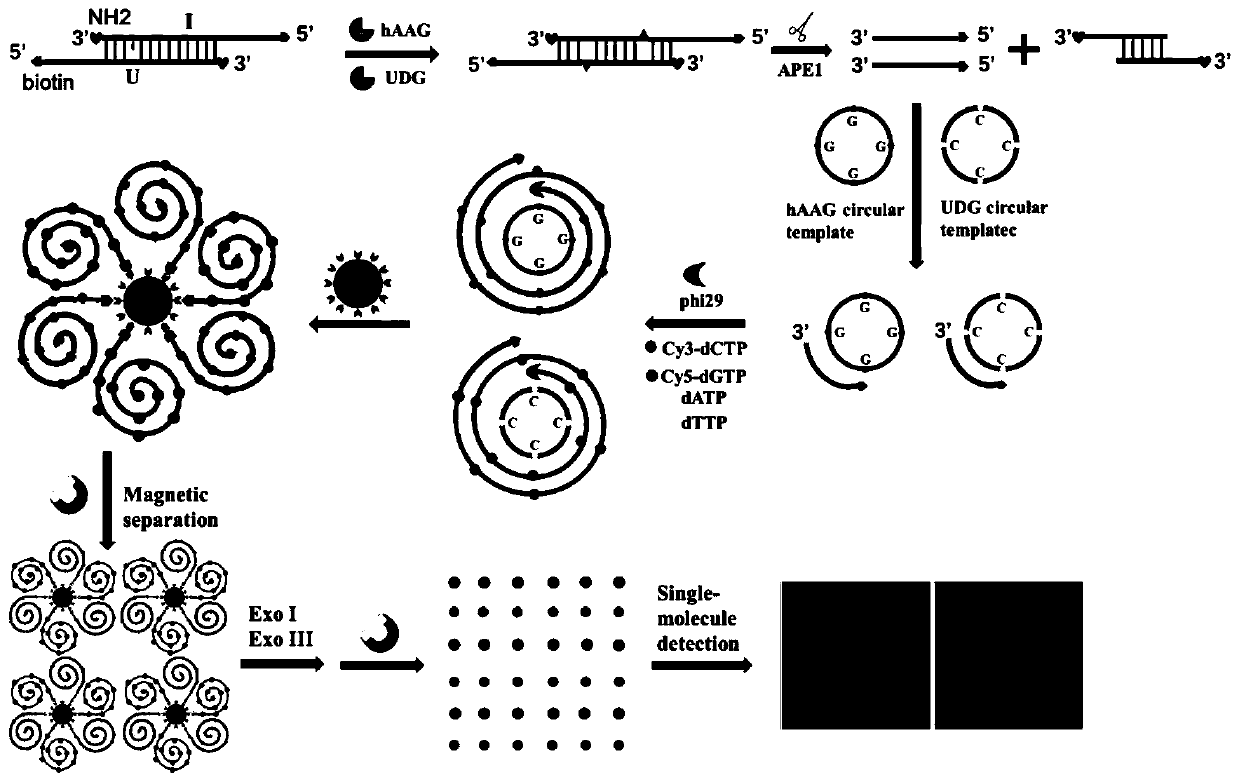
[0086] 1. Preparation of Reagents
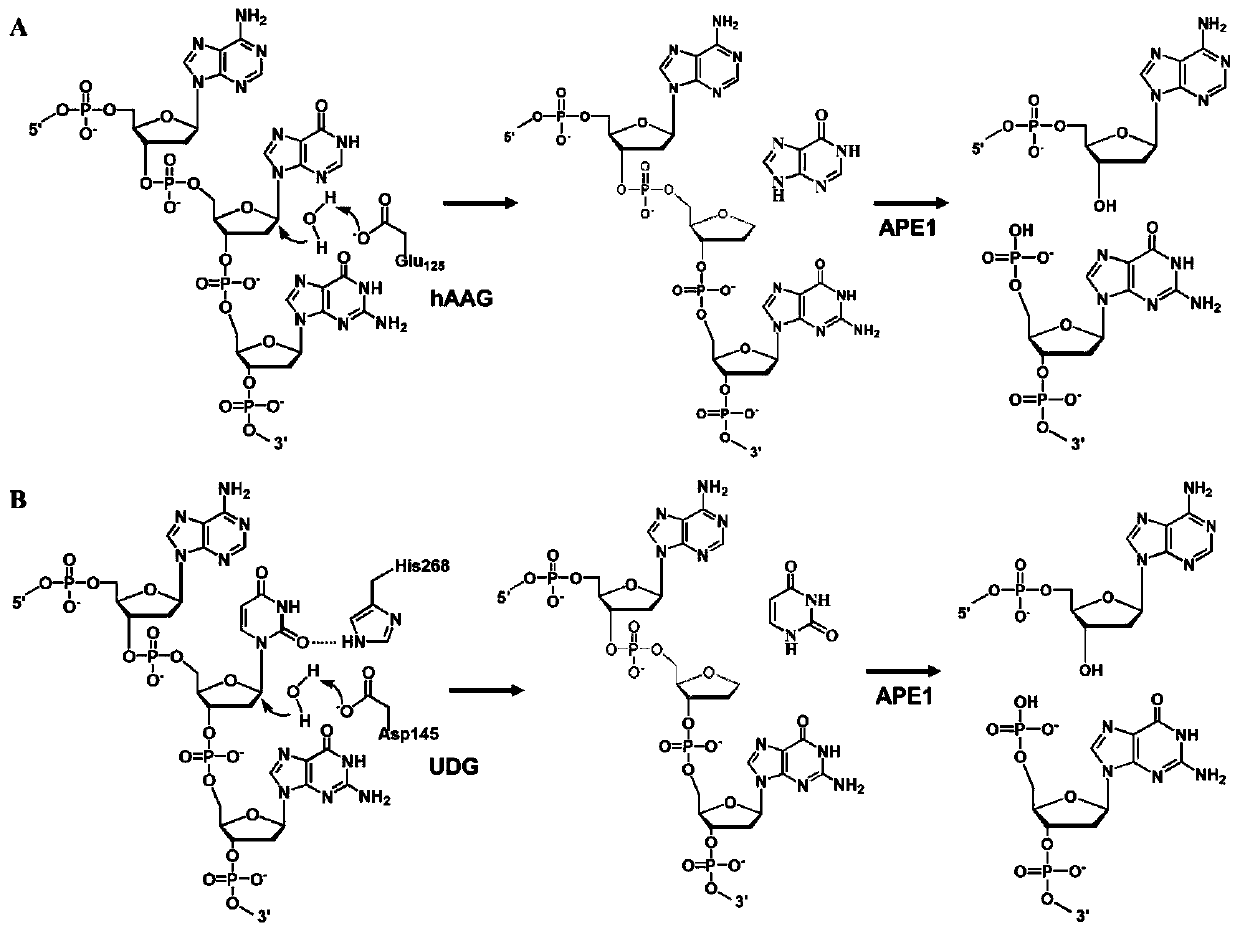
[0087] Preparation of bifunctional dsDNA substrates: 1 μM hAAG probe and 1 μM UDG probe were mixed in 10 mM Tris-HCl (pH 8.0), 50 mM Tris-HCl (pH 8.0) NaCl and 1 mM EDTA in annealing buffer, incubate at 95°C for 5 min, and then slowly cool to room temperature to form a bifunctional dsDNA substrate. The obtained bifunctional dsDNA substrate was stored at 4°C for further use.
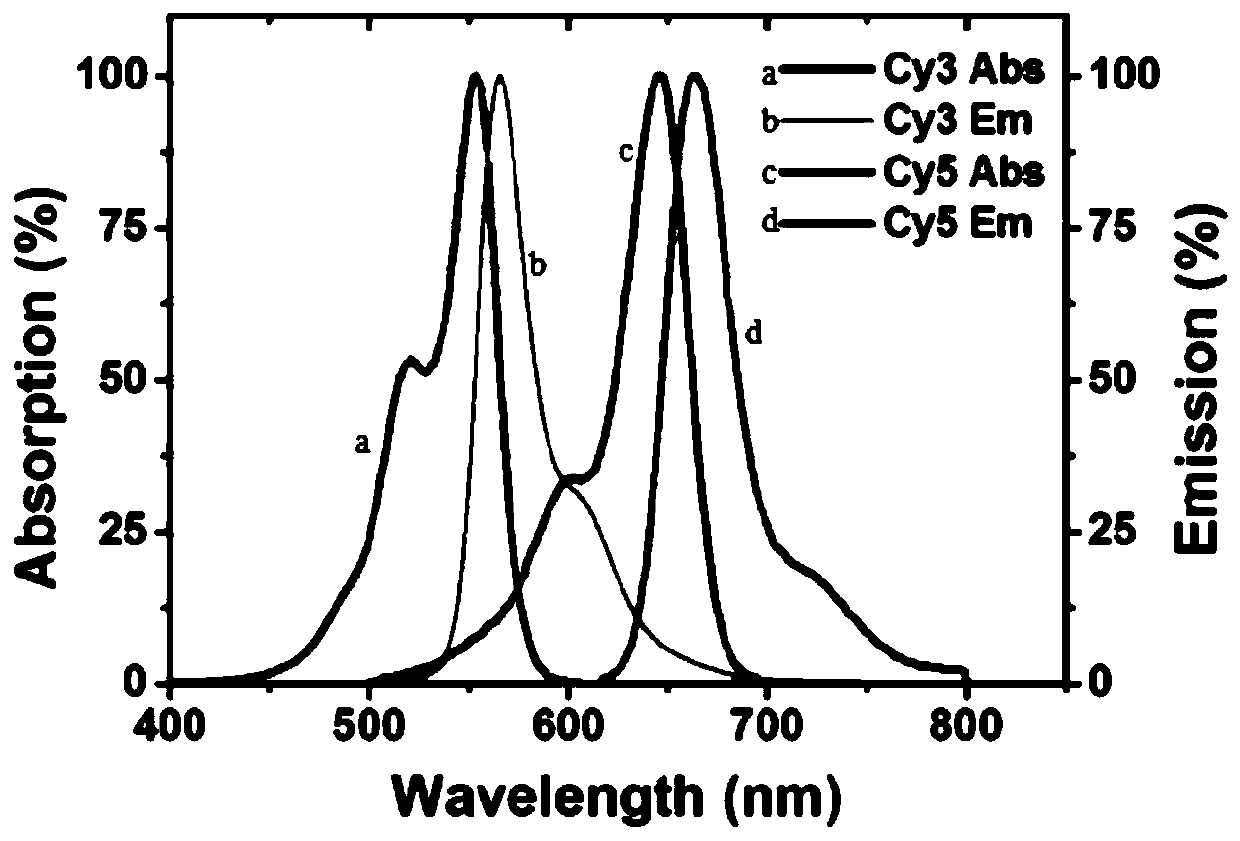
[0088] DNA glycosylase-induced excision reaction and RCA reaction: DNA glycosylase-induced excision reaction was performed in 10 μl reaction solution containing 100 nanomoles per liter of bifunctional dsDNA substrate, 1x NEB buffer 4. 1x UDG reaction buffer, 2U APE1 and different concentrations of hAAG and UDG, react at 37°C for 1 hour. Then combine 50 nmoles per liter of hAAG cyclic template, 50 nmoles per liter of UDG cyclic template, 0.1 mg per ml of BSA, 0.25 mmoles per liter of dATP, 0.25 mmoles per liter of dTTP, 10 Micromoles per liter of Cy3-labeled dCTP (Cy3...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com