Cryptic metabolites and method for activating silent biosynthetic gene clusters in diverse microorganisms
A technology of metabolites and microorganisms, applied in the field of genetics, can solve the problems of slowing down the progress and throughput of natural product discovery, and the method of identifying recessive metabolites has yet to be developed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0060] Unless otherwise defined above, all technical and scientific terms used herein have the same meaning as commonly understood by one of ordinary skill in the art to which this invention belongs. Where a term is provided in the singular, the inventors also contemplate the plural of that term.
[0061] The singular forms "a", "an" and "the" include plural forms unless the context clearly dictates otherwise.
[0062] The terms "comprise" and "comprising" are used in an inclusive, open sense meaning that additional elements may be included.
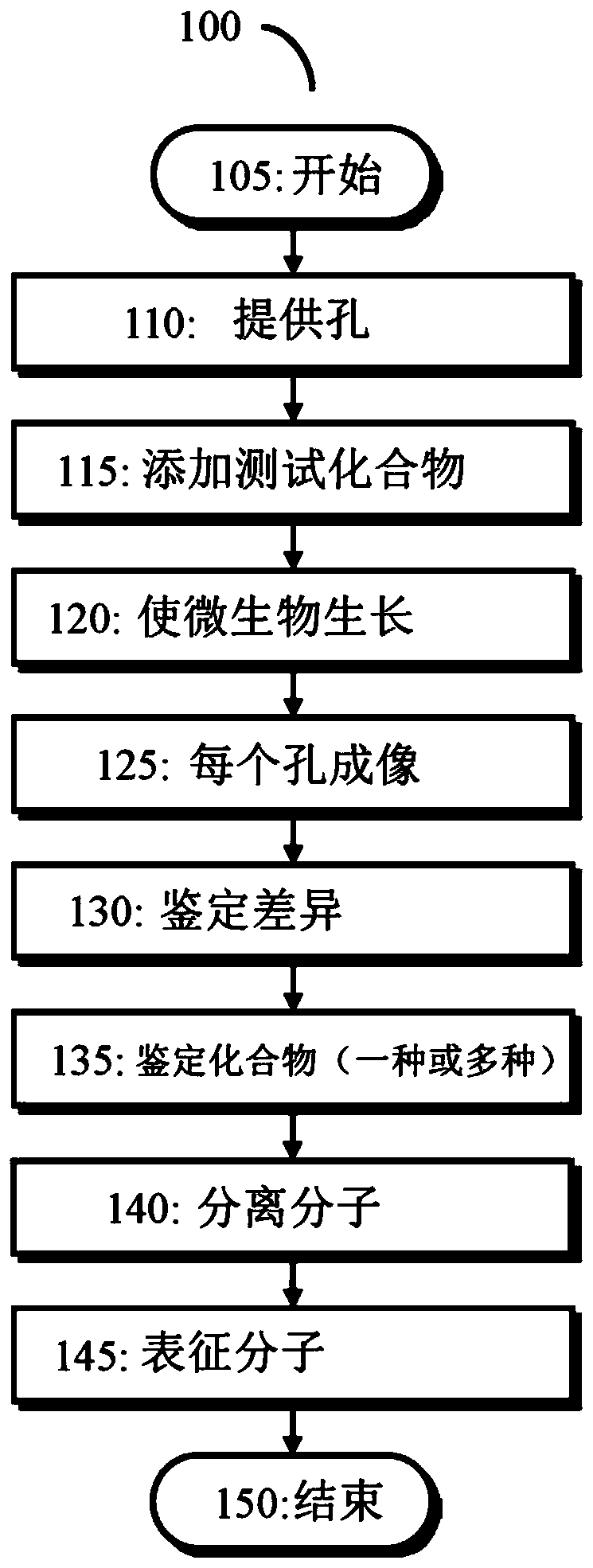
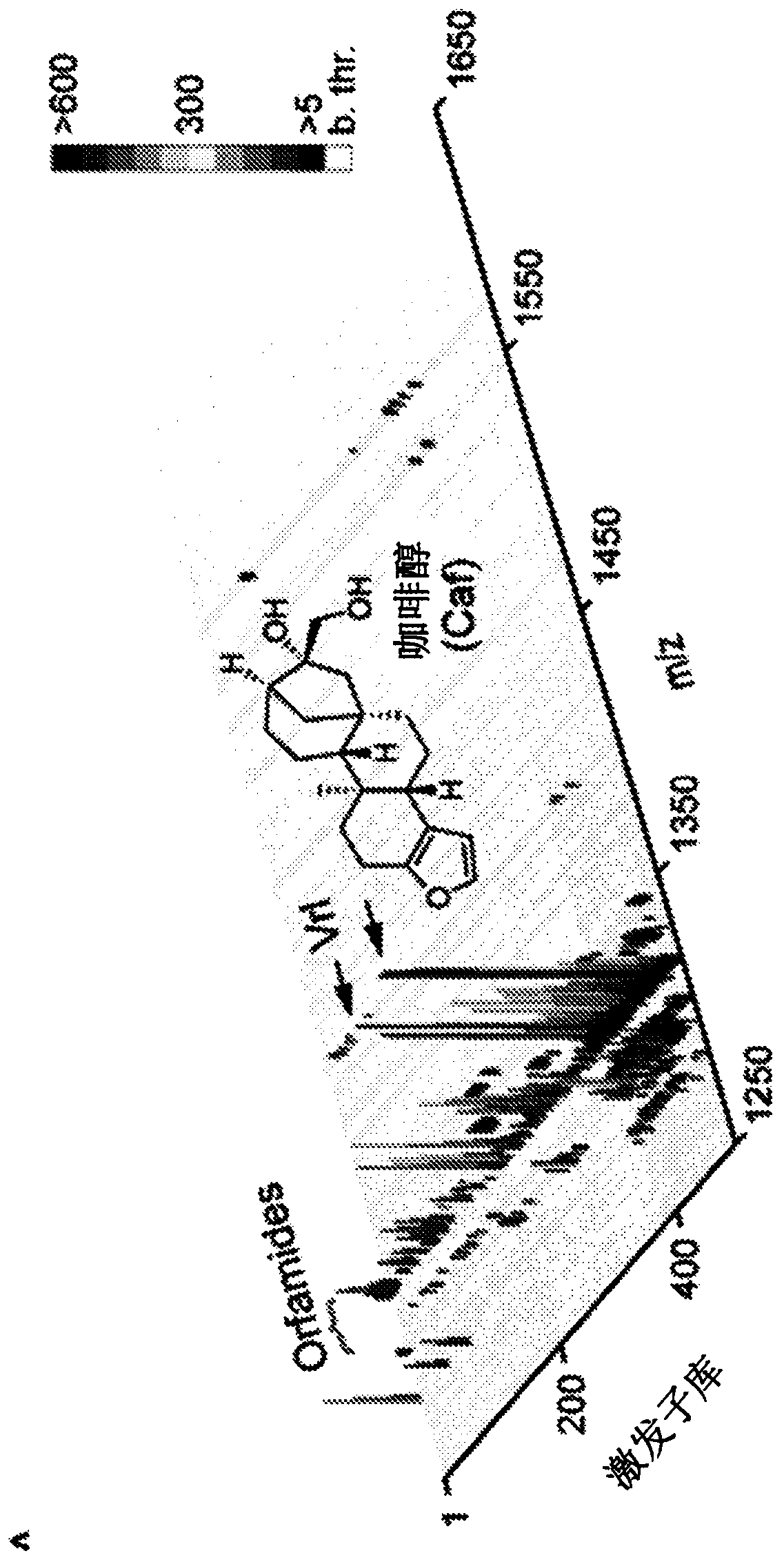
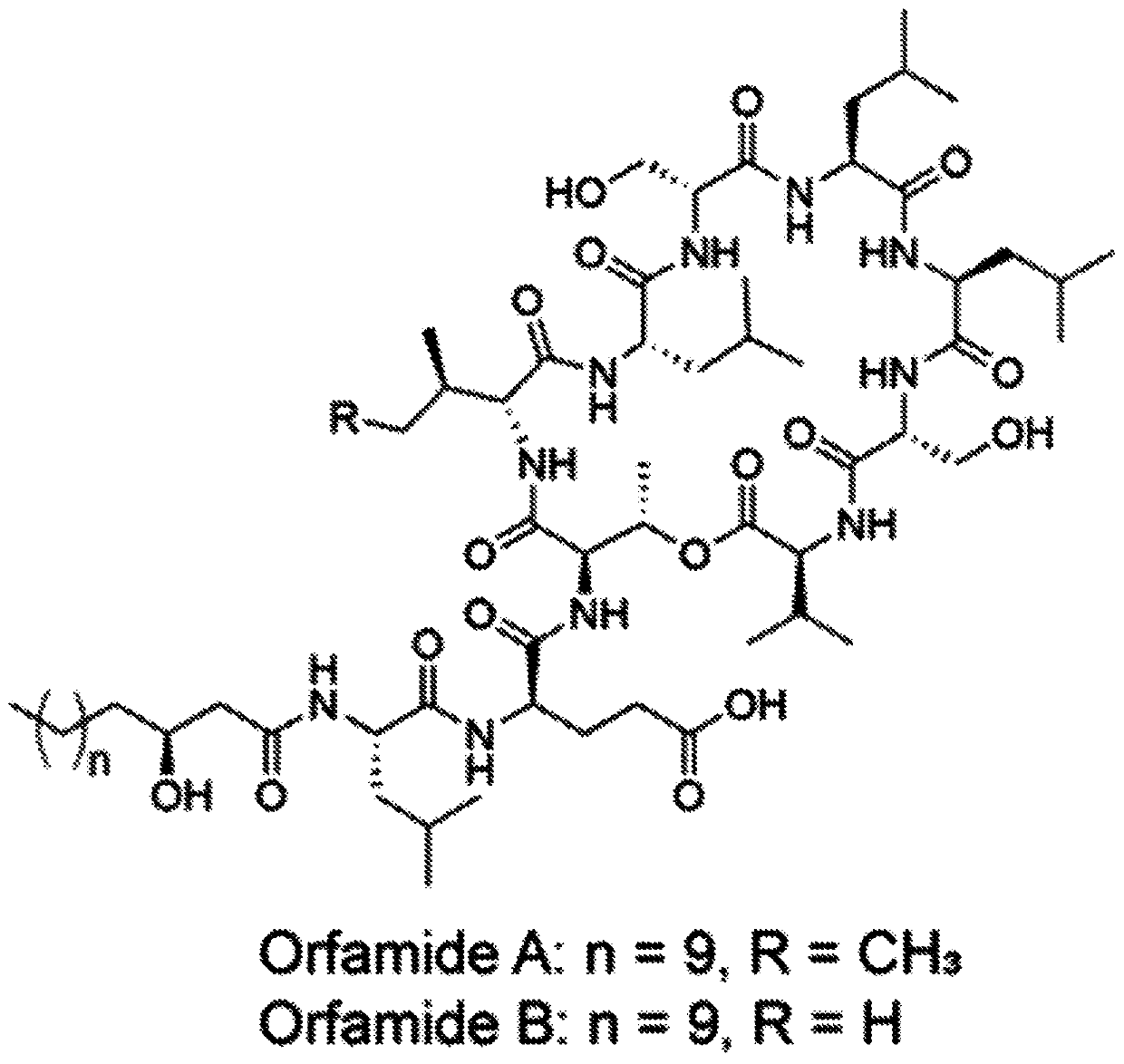
[0063] An inexpensive and non-labour-intensive method for rapid excitation of recessive metabolites is disclosed.
[0064] Silent BGCs are a treasure trove of new secondary metabolites. Advances in DNA technology and widespread genome sequencing have compiled a vast database of genomes that needs to be mined to harvest decades of innovation. The ideal way to do this would be without having to challenge the genetic or cloning procedure...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com