Method for separating trophoblastic cells
A technology of trophoblast cells and antibodies, applied in the field of cell separation, can solve the problems of low acceptance by pregnant women, false positives, long analysis time, etc., and achieve the effect of low risk of infection and abortion, and early sampling time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] 1. Antibody selection
[0024] According to the specific antigens expressed on the surface of various types of trophoblast cells obtained by searching the literature, the antigens are shown in Table 1:
[0025] Table 1: Antigens obtained by screening
[0026]
[0027]
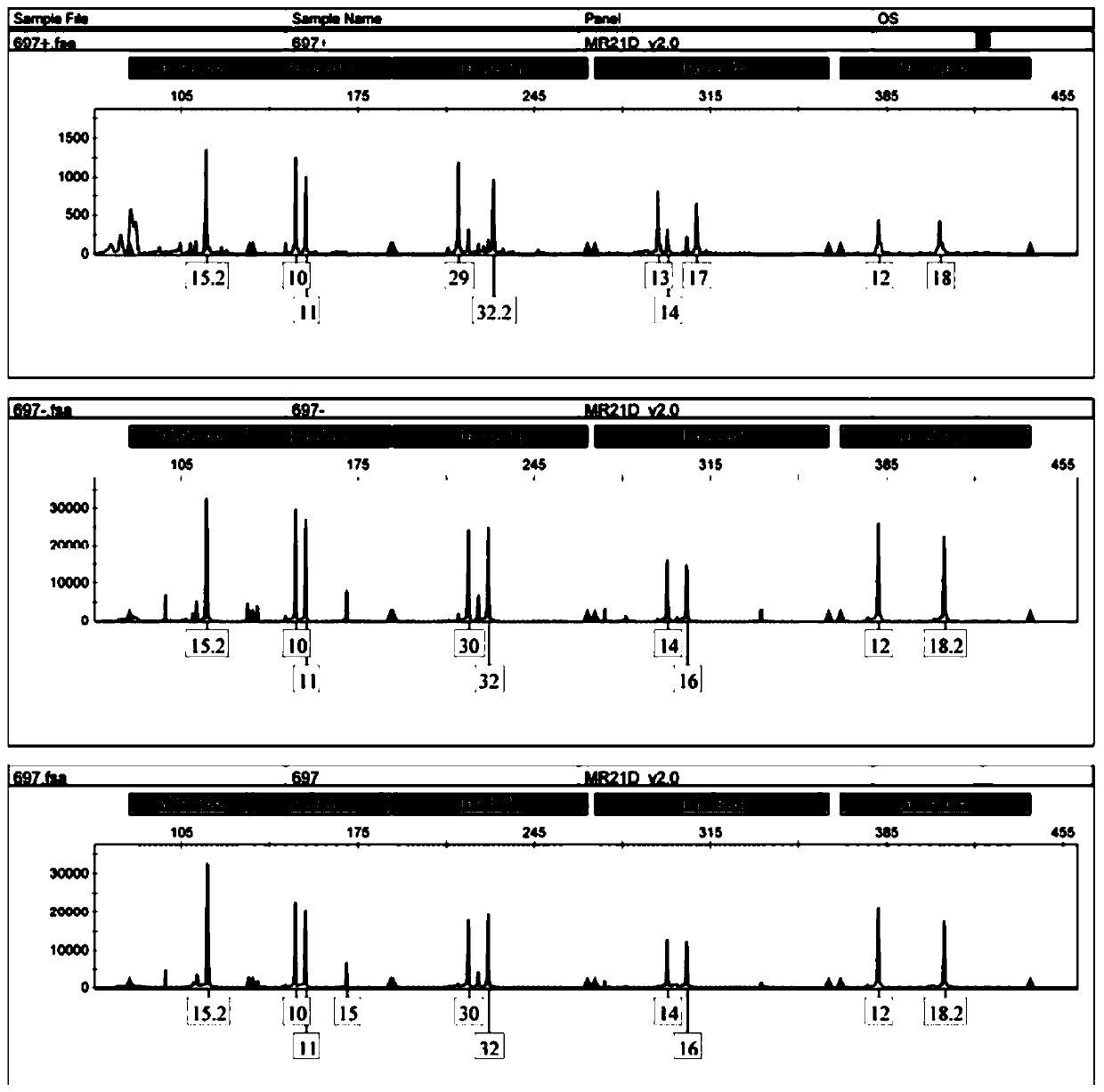
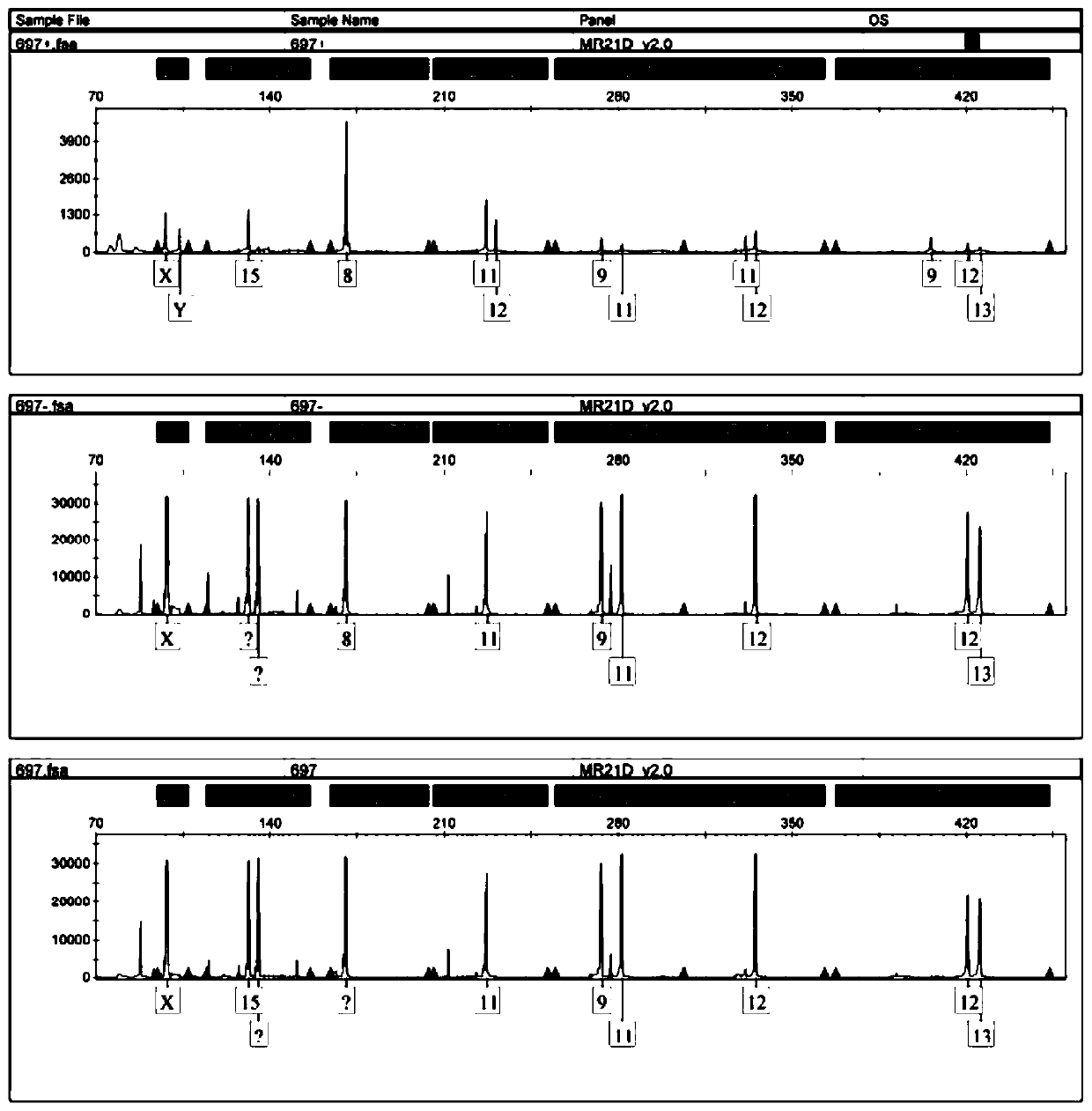
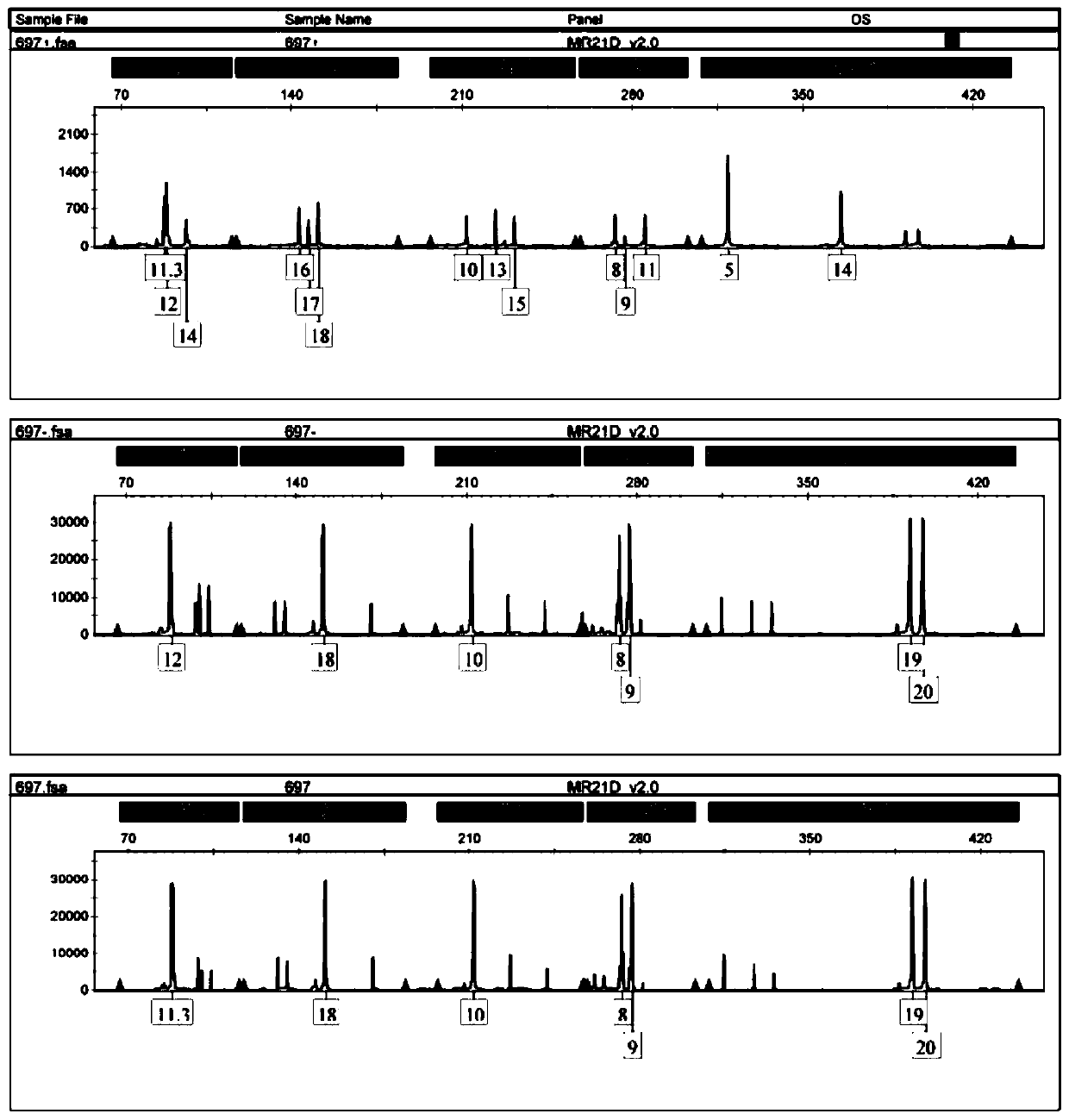
[0028] Cervical trophoblast cells are divided into cytotrophoblast cells, intermediate trophoblast cells, and syncytiotrophoblast cells; there is no uniform international standard for the identification of trophoblast cells, but the more recognized molecular markers are cytokeratin (cytokeratin, CK), vimentin, human chorionic gonadotropin (β-hCG), human placental lactogen (hPL), matrix metalloproteinase 9 (MMP9), etc. HLA-G molecules are expressed on embryos, extravillous trophoblast cells at the maternal-fetal interface, amniotic endothelial cells, and fetal vessel wall endothelial cells in the placenta during the entire pregnancy; trophoblast cells are cells with secretory characteristics and ca...
Embodiment 2
[0061] 1. DNA extraction from cervical exfoliated cells of pregnant women:
[0062] (1) Centrifuge 200 ul of cells from the 14th step of cell sorting (remaining cells in which trophoblast cells have been removed from cervical exfoliated cells) and sorted trophoblast cells at 12000 rpm for 3 min.
[0063] (2) Discard the supernatant, add 200ul solution P to resuspend the precipitate.
[0064] (3) Add 20ul of proteinase K and 200ul of solution L, and mix well.
[0065] (4) Incubate at 56°C for 20 minutes, and mix by inverting.
[0066] (5) Add 200 ul of absolute ethanol, mix thoroughly, transfer to an adsorption column, centrifuge at 10,000 rpm for 1 min, and discard the waste liquid in the collection tube.
[0067] (6) Add 500ul of W1, centrifuge at 10000rpm for 1min, discard the waste liquid in the collection tube.
[0068] (7) Add 500ul of W2, centrifuge at 10000rpm for 1min, discard the waste liquid in the collection tube.
[0069] (8) Add 500ul of W2, centrifuge at 1000...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com