Genome integration method and application
A genome and integrated carrier technology, applied in the field of genetic engineering and biology, can solve the problems of reduced editing efficiency and achieve the effect of simple integration method, simplified time-consuming and labor-intensive, and stable genetics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1 Large fragment genome integration technology system construction
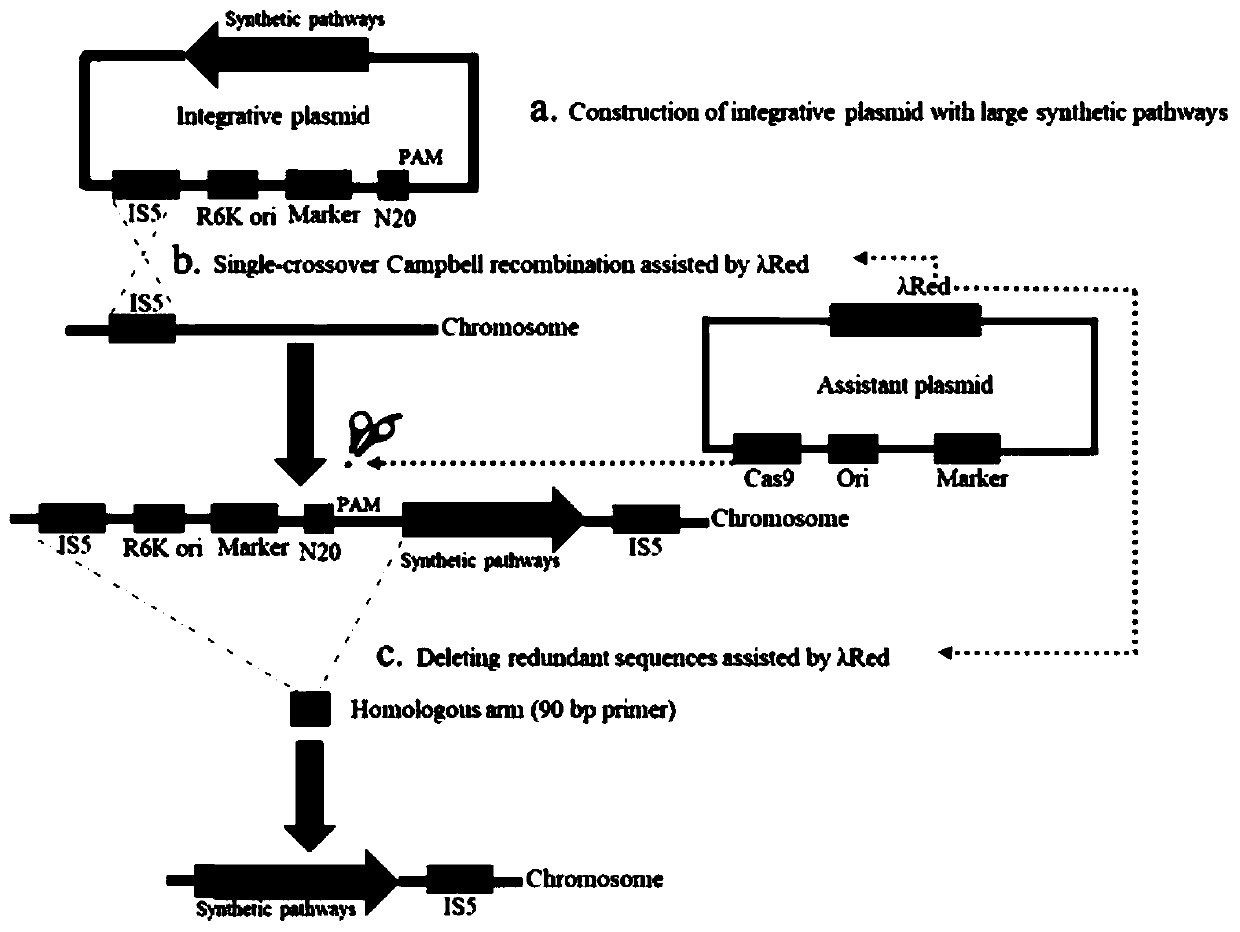
[0029] Genome integration technology process, see figure 1 , first construct the integrated plasmid containing IS5 sequence, R6K replicon, chloramphenicol resistance gene, target exogenous metabolic pathway, N20 recognition site ( figure 2 ). This system mainly relies on plasmids expressing λ-Red recombinant protein and CRISPR / Cas9 system. With the assistance of the λ-Red recombination protein, the integration plasmid was integrated into the E. coli genome through Campbell-type recombination. Then under the mediation of CRISPR / Cas9, redundant sequences such as IS5 sequence, R6K replicon, and chloramphenicol resistance gene were deleted.
Embodiment 2
[0030] Embodiment 2 different module promoter selection
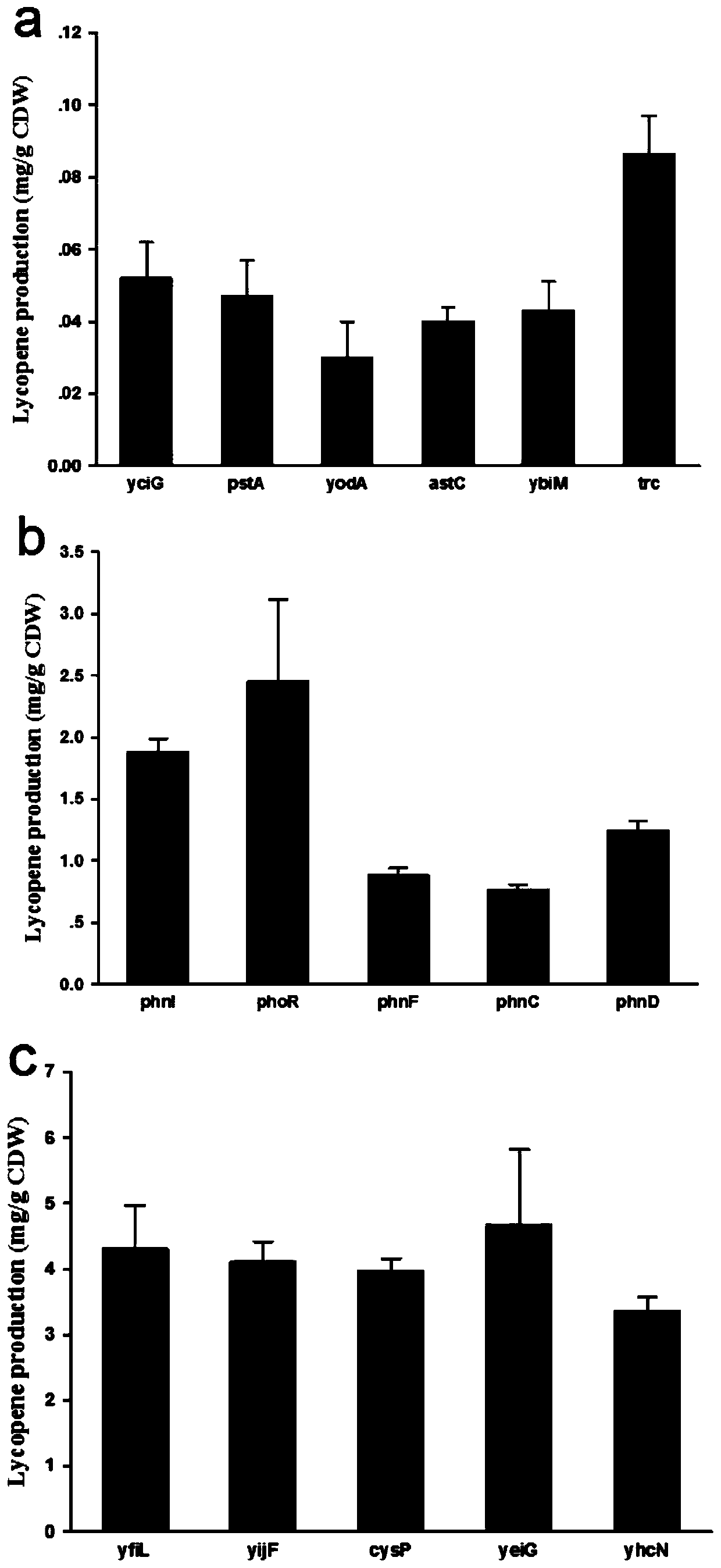
[0031] 1. Screening of promoters of lycopene synthesis pathway
[0032] This example first uses the pET-30a-trc vector preserved in the laboratory (Su B, Zhang Z, Wu M, Lin J, YangL: Construction of plasmamid-free Escherichia coli for the production of arabitol-free xylitol from corncob hemicellulosic hydrolysate.Sci Rep 2016,6:26567.) as a template, with primers trc-30a-P1 / trc-30a-P2 to amplify the plasmid module, with primers crtI-P1 / crtI-P2, crtE-P1 / crtE-P2, crtB-P1 / crtB-P2 respectively amplifies lycopene synthesis genes crtI, crtE, crtB (crtI, crtE, crtB gene sequence has been disclosed in the patent application number CN 109943492A, the name of the invention: a Chinese patent application for a recombinant yeast strain and its application ), using a recombinant kit to construct the vector pET-trc-IEB. Subsequently, the laboratory preserved pCDFDuet-1 (Su B, Zhang Z, Wu M, Lin J, Yang L: Construction of plasma-free ...
Embodiment 3
[0054] Example 3 Large Fragment Genome Integration
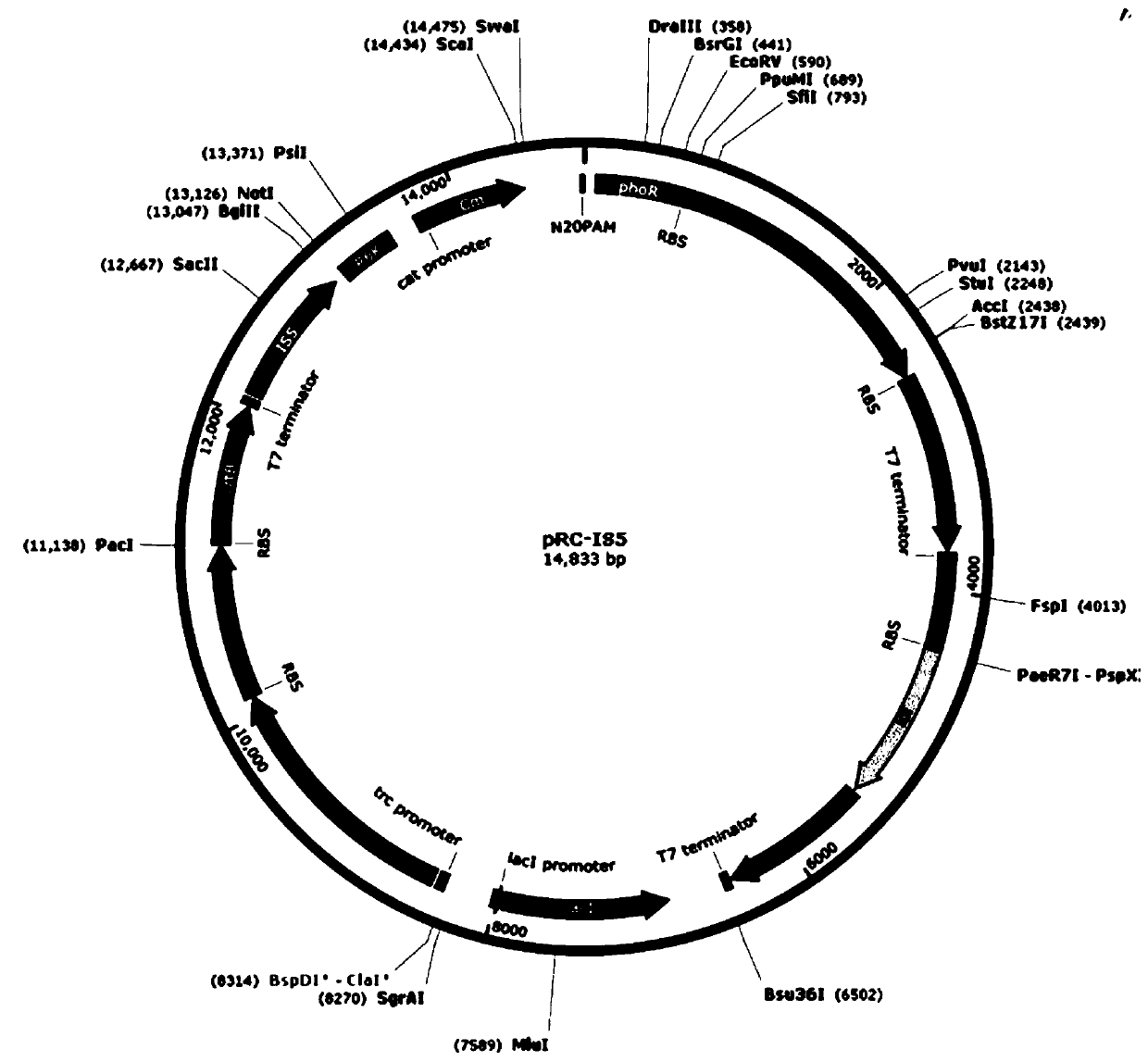
[0055]Using pACYC-phoR (pACYC-phoR-dxs-dxr-yciG-IEB) as a template, primers phoR-dxs-dxr-P1-3, phoR-dxs-dxr-P1-2, phoR-dxs-dxr-P1 / phoR-dxs-dxr-P2 amplifies the phoR-dxs-dxr module; using pACYC-yejG (pACYC-yejG-idi-crtE-yciG-IEB) as a template, using primers yejG-idi-crtE-P1 / yejG-idi- crtE-P2 amplifies the yejg-idi-crtE module; uses pET-trc-IEB as a template, and uses primers IEB-P1 / IEB-P2 to amplify the lacI-trc-IEB module; uses pRC43 (Su B, Zhang Z, Wu M ,Lin J, Yang L:Construction ofplasmid-free Escherichia coli for the production of arabitol-free xylitol from corncob hemicellulosic hydrolysate.Sci Rep 2016,6:26567.) as a template, using primers IS5-R6K-CM-P1 / IS5-R6K -CM-P2, IS5-R6K-CM-P2-2 amplify the IS5-R6K-Cm module, and use the recombination kit to construct the recombinant plasmid pRC-IS5( figure 2 ), the recombinant plasmid pRC-IS5 contains IS5 sequence, R6K replicon, chloramphenicol resistance gene, target exog...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com