Method for tracing blood lineage sources of buffaloes and carrying out genome matching based on whole-genome SNP information
A whole-genome and genome-wide technology, applied in the field of animal breeding, can solve problems such as the lack of detailed buffalo genetic relationships, unreliable division methods, and lack of pedigree information, and achieve the effects of reducing the probability of inbreeding, improving the production performance of offspring, and accurately tracing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
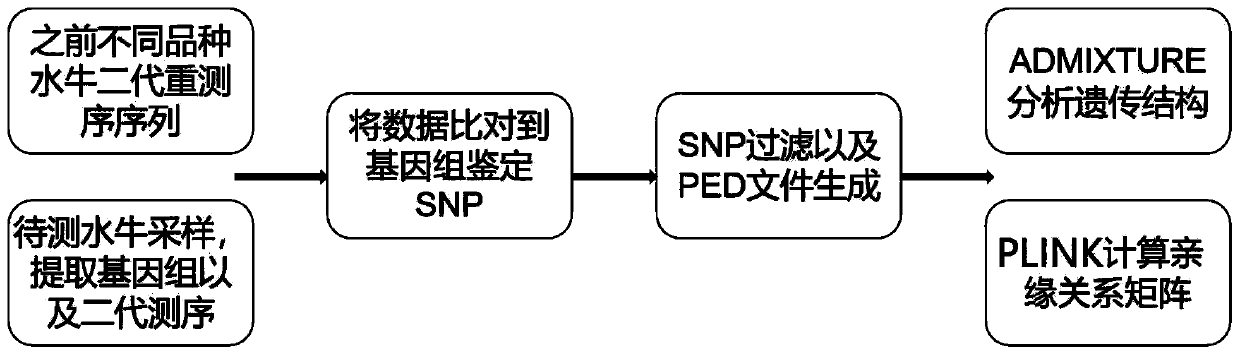
[0045] Example 1 Lineage traceability analysis and genome selection based on genomic SNP data
[0046] Experimental material: Blood samples from 4 test hybrid buffaloes. All buffaloes have been subjected to genome SNP genotype mining through Illumina next-generation data sequencing technology.
[0047] Specific steps are as follows:
[0048] (1) Extract blood sample DNA from all buffaloes and sequence them on the IlluminaHiSeq high-throughput sequencing platform; download the second-generation sequencing data of the world's major buffalo breeds from the BIG Sub database (BIG Sub: CRA001463), and obtain the genome SNP genes type information.
[0049] (2) SNP quality control: The raw data obtained in step (1) were quality controlled by Fastqc and Trimmomatic software, and the processed fragments were compared to the reference genomes of swamp buffalo and river buffalo (GWHAAJZ00000000 and GWHAAKA00000000) by BWA software , after base quality correction, SNP typing was perform...
Embodiment 2
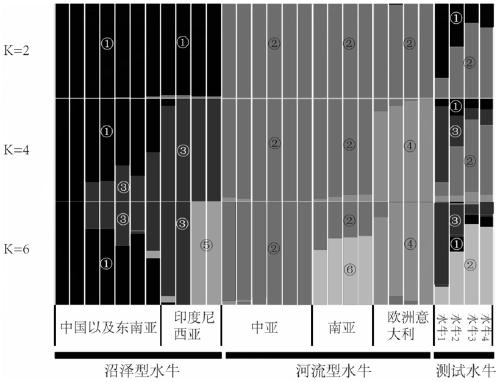
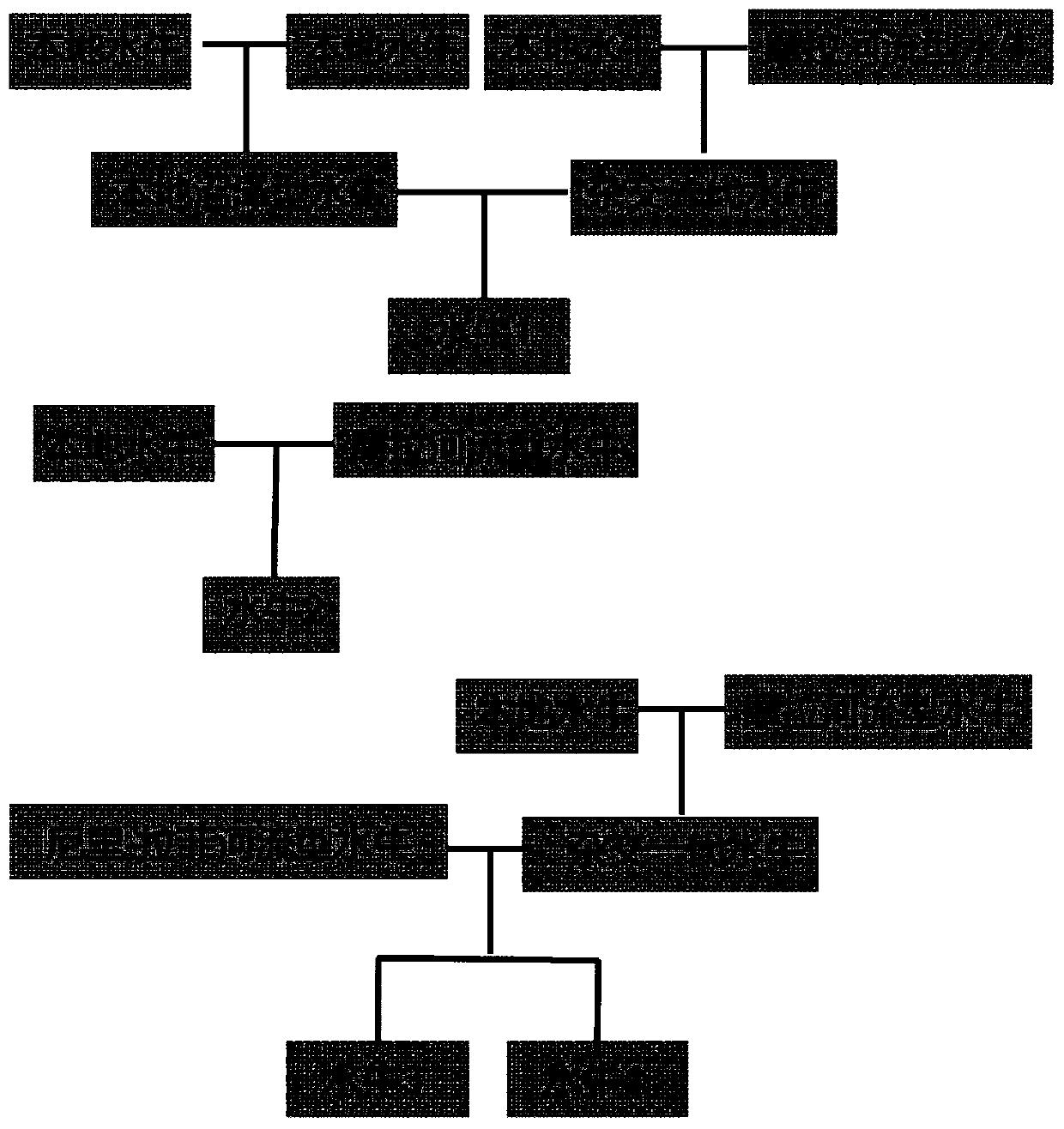
[0054] Example 2 The comparison between the new lineage tracing method and the pedigree results
[0055] Experimental material: Test hybrid buffalo map
[0056] Based on the ancestral component analysis of the Bayesian method and the buffalo SNP information collected in Example 1, use ADMIXTURE to analyze the genetic structure of the buffalo population. The preset subgroup number (K value) is 2-8, and the SNP of each individual is carried out. Bayesian inference subgroups, and finally calculate the proportion of different components of all SNPs for each individual.
[0057] When the number of subgroups (K value) is 2, the structure at this time is the composition of swamp buffalo and river buffalo. ADMIXTURE will calculate the proportion of these two parts of all individuals, and preliminarily deduce it based on the proportion of the two parts in the hybrid buffalo For the number of hybrid generations, when the proportion of ingredients is 40-60%, it is the first generation...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com