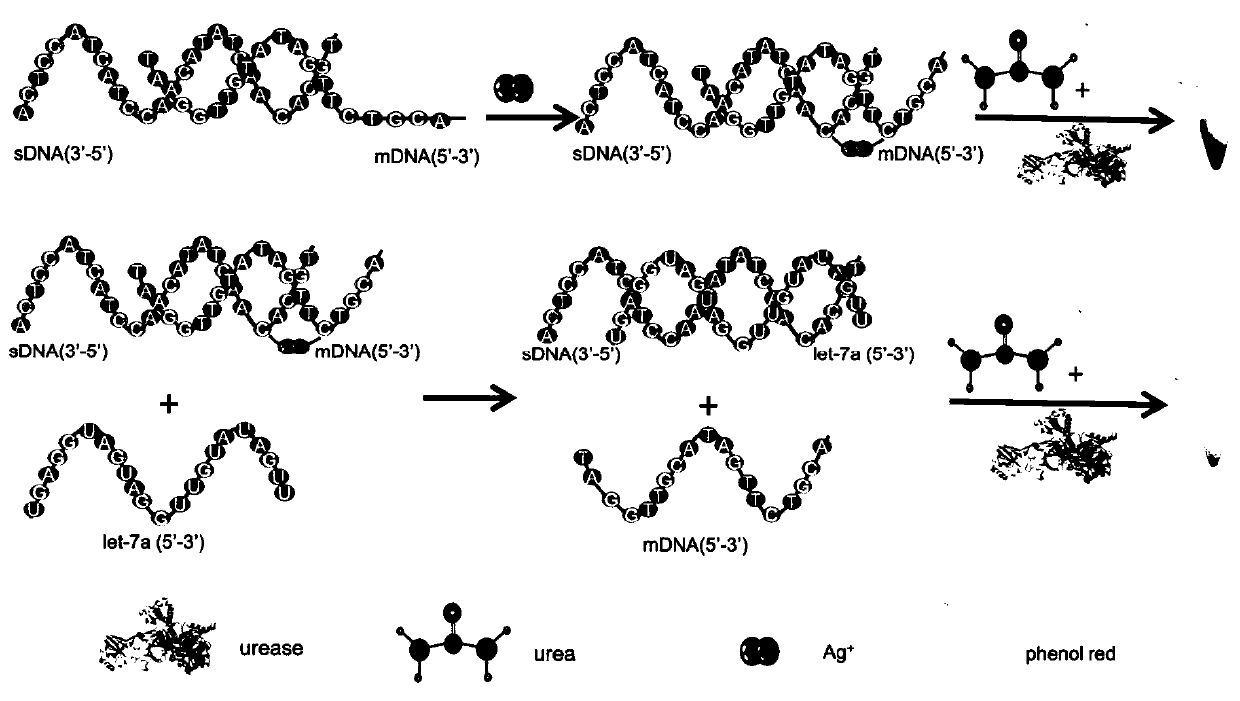
Urease based microRNA detection kit
A detection kit, the technology of the kit, which is applied in the determination/inspection of microorganisms, the measurement of color/spectral properties, and the material analysis by observing the influence on chemical indicators, can solve the problems of nucleic acid molecule detection that have not been seen before, To achieve the effect of simplified operation, convenient detection and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Embodiment 1 prepares double-stranded DNA solution
[0049] In this embodiment, a solution of double-stranded DNA is prepared, and the double-stranded DNA plays a role of molecular recognition in the process of detecting miRNA.
[0050] First, the substrate strand and the mismatched strand are synthesized, and their sequences are shown in Table 1.
[0051] The toehold structure refers to the single-stranded part of the double-stranded DNA that cannot be paired at the end. In Table 1, the underlined part of the substrate strand / mismatched strand sequence is the toehold structure, and the sequence numbers of the strands are all defined by the length of the toehold. Substrate strands and mismatched strands with the same sequence number are paired and used in conjunction with each other, for example, the substrate strand with sequence number 3nt (sSub-3nt) is used in conjunction with the mismatched strand with sequence number 3nt (mSub-3nt).
[0052] The bases of the toeh...
Embodiment 2
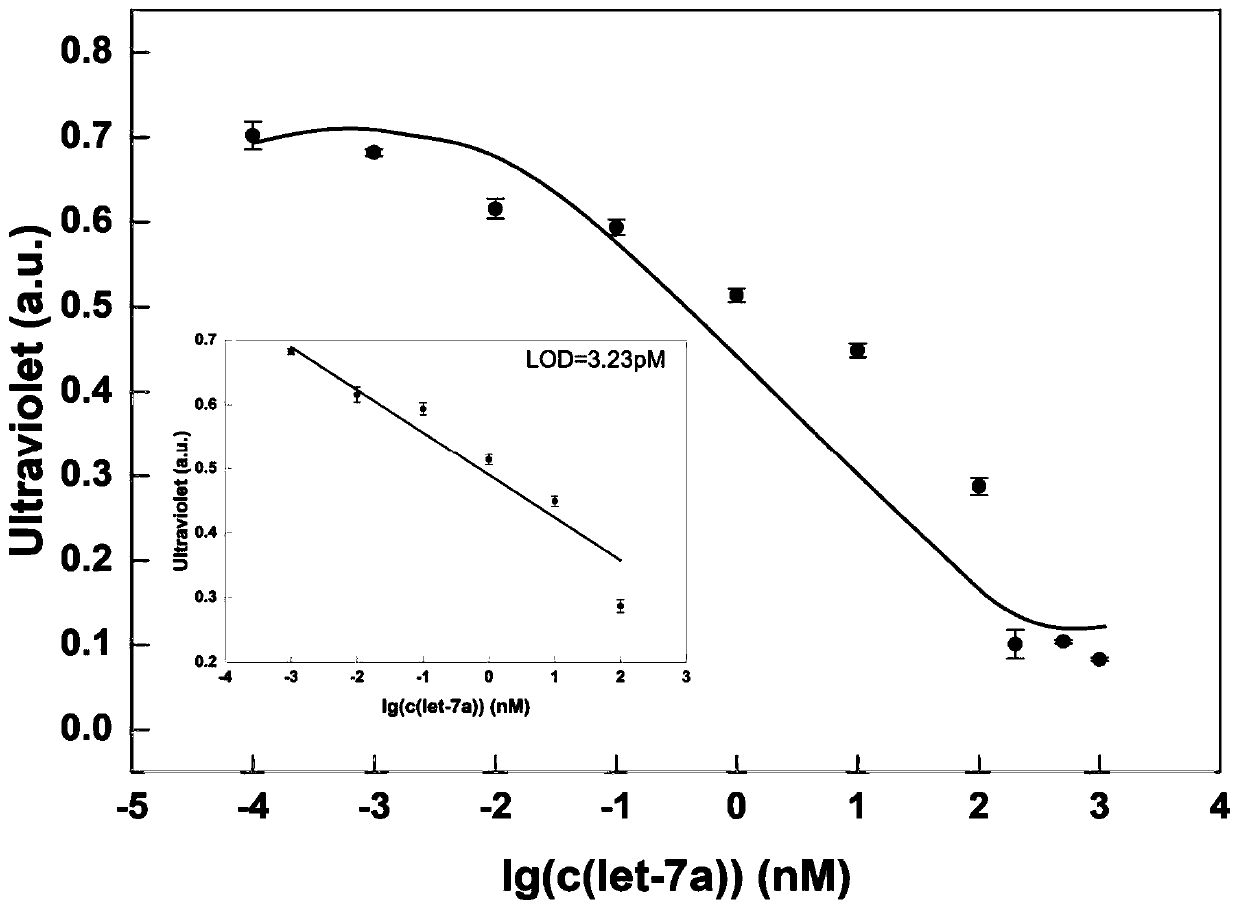
[0059] Embodiment 2 draws standard curve
[0060] In the present embodiment, the standard solution curve is drawn, and the steps are as follows:
[0061] ① Take 30 μL of the double-stranded DNA solution containing the C-C structure prepared in Example 1, add 10 μL of let-7a solution with a concentration of 0 nM to it, and place it at room temperature for 30 minutes to carry out the displacement reaction. Then 6 μL of urease with a concentration of 100 nM was added, mixed with 4 μL of ultrapure water to maintain a total volume of 50 μL, and then incubated at room temperature for 30 min to obtain a reaction mixture.
[0062] ② Take 10 μL of 2.5 mM phenol red solution and 10 μL of 5 M urea aqueous solution at room temperature and add them to 30 μL of ultrapure water to obtain 50 μL of urea-phenol red mixed solution.
[0063] ③ Mix the solutions prepared in ① and ②, react at room temperature for 20 minutes, measure the ultraviolet absorption of the reaction mixture at a wavelengt...
Embodiment 3
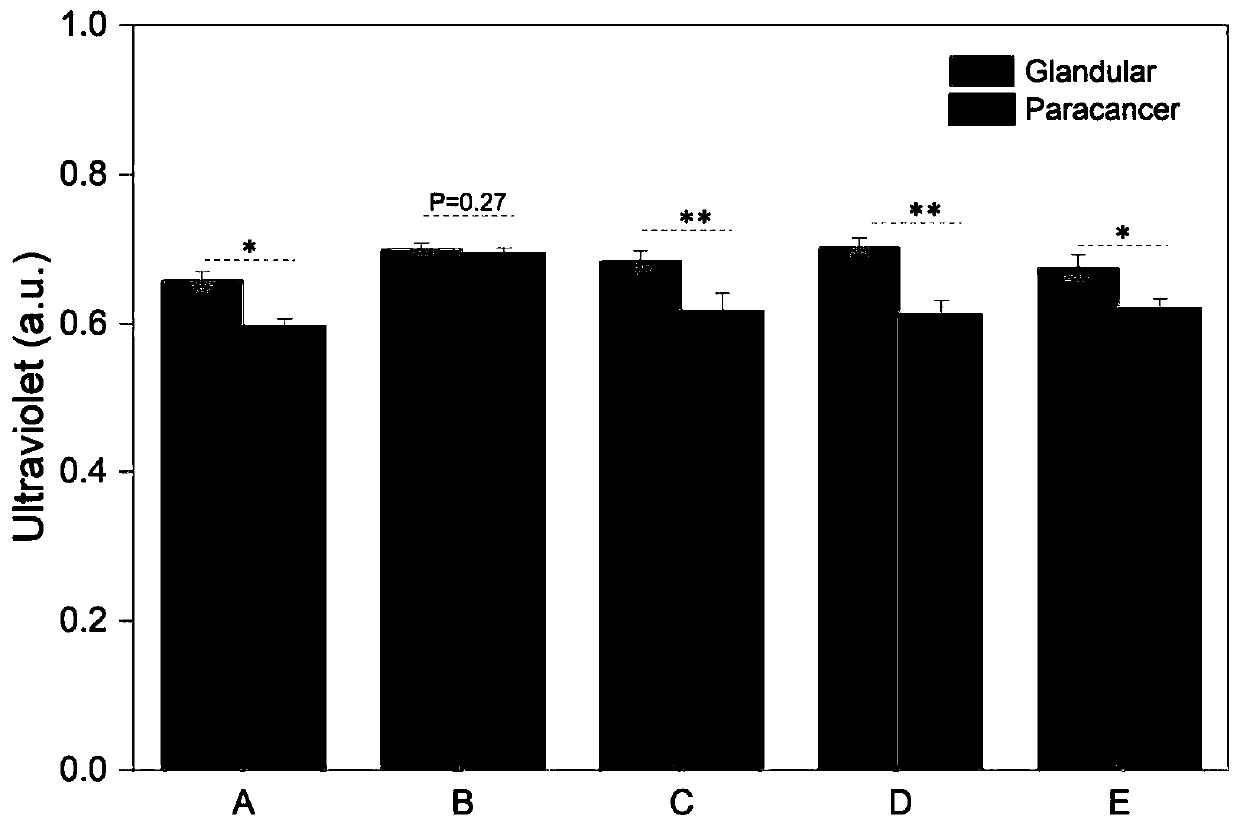
[0066] Embodiment 3 clinical detection
[0067] In this example, the concentration of let-7a in cancer tissues and paracancerous tissues from 5 lung cancer patients was detected, and the significant difference was analyzed. The steps are as follows:
[0068] (1) The cancer tissues and paracancerous tissues of 5 lung cancer patients were collected, and recorded as 1#~5# in turn, the cancer tissues were recorded as C, and the paracancerous tissues were recorded as A, and the total RNA of 1#~5# tissues was extracted, Make RNA aqueous solution.
[0069] (2) Let-7a detection in RNA aqueous solution
[0070] ①Take two parts (30 μL) of the double-stranded DNA solution containing the C-C structure prepared in Example 1, add 1#A and 1#C sample solutions to it in turn, and place it at room temperature for 30 minutes to carry out the displacement reaction. Then 6 μL of urease with a concentration of 100 nM was added, mixed with 4 μL of ultrapure water to maintain a total volume of 50 μ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com