Method for reprogramming ITNK cells based on CRISPR/Cas9
A reprogramming and cell technology, applied in the field of gene editing, can solve the problems of high toxicity and low efficiency of ITNK cells, and achieve the effects of strong killing ability, good in vitro expansion ability, and strong in vitro expansion ability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] This embodiment provides a method for reprogramming ITNK cells, the specific steps are as follows:
[0061] 1. Gene sequences of sgRNA, cr-RNA and tracr-RNA.
[0062] (1) According to the Bcl11b gene, design three sgRNA targeting the gene, that is, SEQ ID NO.1~3, and send the sequence to the sequence synthesis company to synthesize the sequence, and connect the three sgRNAs to the PX458-gBCL11b vector containing the T7 promoter Above, the successful construction of the vector was verified by sequencing, and the specific sequence of the sgRNA used was as follows:
[0063] sgRNA1: gaccctgacctgctcacctg (SEQ ID NO.1)
[0064] sgRNA2: gaccatgaactgctcacttg (SEQ ID NO.2)
[0065] sgRNA3: gaagcagtgtggcggcagct (SEQ ID NO. 3);
[0066] (2) Through mMESSAGE mMACHINE TM The T7 Transcription Kit kit is used to obtain the mRNA product of the gRNA targeting the gene in vitro;
[0067] (3) The gene sequences of crRNA and tracrRNA used in the present embodiment are as follows:
...
Embodiment 2
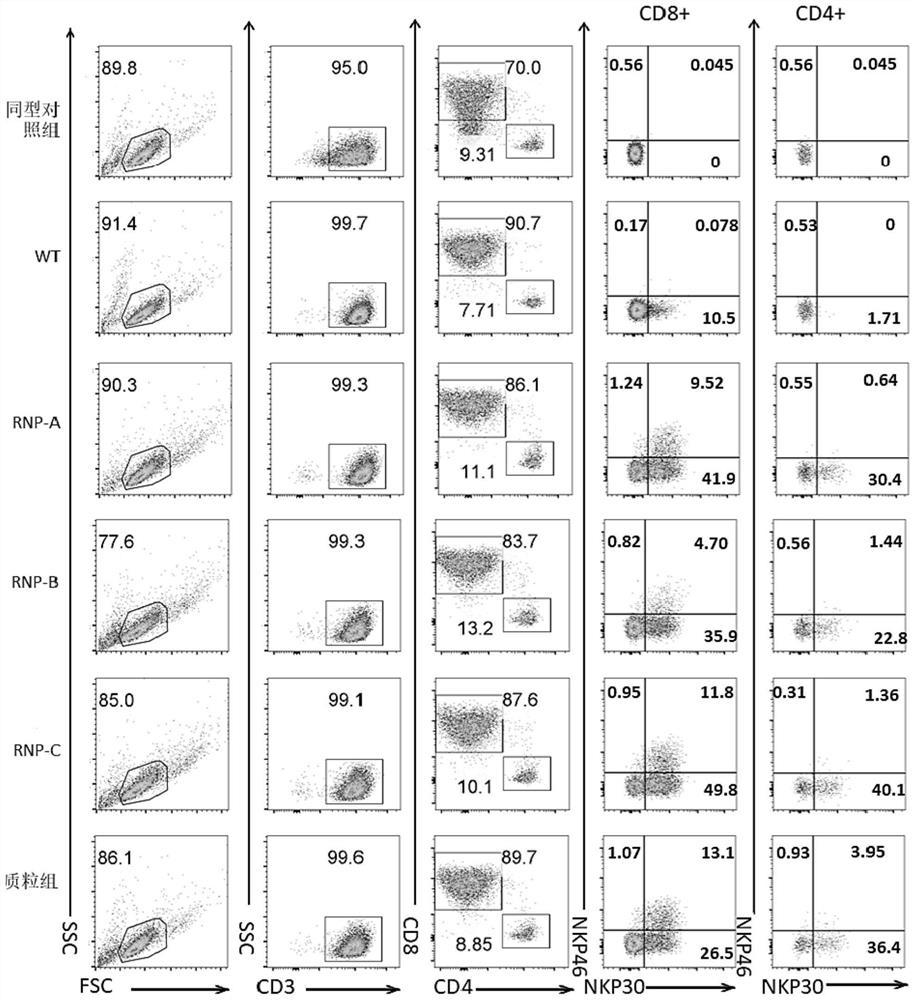
[0082] In this example, the growth state and phenotype of cells in each experimental group in Example 1 were monitored and compared.
[0083] On the 9th day after electroporation, subgroups expressing both T cell marker CD3 and NK cell markers NKp46 and NKp30 appeared in the electroporation RNP group (RNP-A, RNP-B and RNP-C group) and the plasmid group.
[0084] Experimental results such as figure 1 As shown, in the CD8 positive cell population, the positive ratio of NKp30 in the plasmid group was only 39.6% (26.5%+13.1%), while the ratio of NKp30 in the electroporation RNP-A, B and C groups were 51.42%, 40.6% and 61.6% respectively , the proportion of NKp46 had no significant difference;
[0085] In the CD4-positive cell population, the proportion of NKp30 in the plasmid group was 40.35%, and that in the RNP-C group was 41.46%, which were similar.
[0086] Based on the above results, compared with direct electroporation of CRISPR / CAS9 plasmids, the proportion of T cells wit...
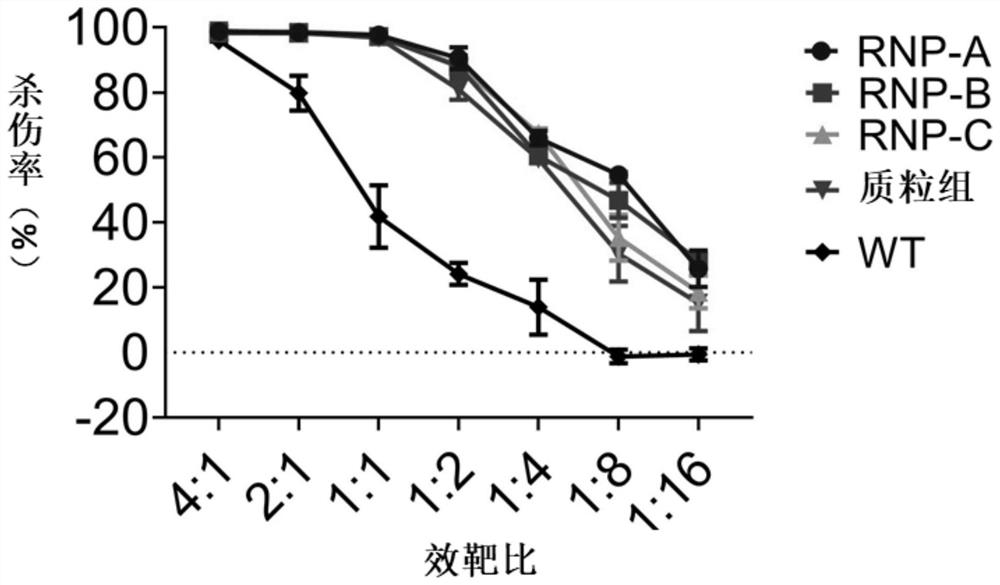
Embodiment 3
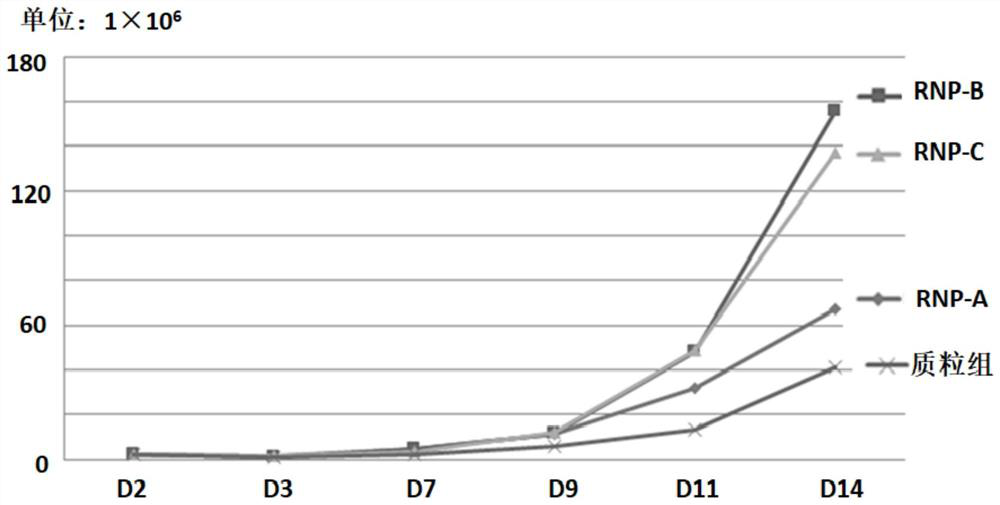
[0088] This example is used to compare the in vitro expansion ability of reprogrammed ITNK cells obtained by electroporation of RNP complexes and direct electroporation of plasmids.
[0089] After the electroporation was completed and the fresh medium was replaced, the growth status of the cells was monitored regularly; the cells were counted with trypan blue every other day to obtain figure 2 The in vitro expansion curves of each group of cells shown and the number of cells in different groups recorded in Table 1.
[0090] Table 1
[0091] group RNP-A group RNP-B group RNP-C group Plasmid set The number of cells in D2 / (×10 6 indivual)
[0092] The above results show that the starting cell number of each group is 2.5×10 6 Taking the day of electroporation as D1, and the day after electroporation (D3), only 0.98×10 cells in the ITNK plasmid group were directly electroporated. 6 , lower than the other three groups of electrotransfer RNP; after the com...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com