A kind of isolated nucleic acid molecule and its use
A technology of nucleic acid molecules and uses, applied in the field of biomedicine, can solve the problems of affecting lipid decomposition, ineffective treatment methods, and unclear specific signs and symptoms of BCD.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0368] 1. Use of CYP4V2 and RdCVF in the preparation of medicaments for treating, alleviating and / or preventing diseases or conditions associated with retinal pigment epithelium (RPE) atrophy.
[0369] 2. The use according to embodiment 1, wherein the disease or condition comprises crystalline retinal degeneration.
[0370] 3. The use according to any one of embodiments 1-2, wherein the CYP4V2 is human CYP4V2.
[0371] 4. The use according to any one of embodiments 1-3, wherein the CYP4V2 comprises the amino acid sequence shown in any one of SEQ ID NO: 76-82.
[0372] 5. The use according to any one of embodiments 1-4, wherein the polynucleotide encoding CYP4V2 comprises the nucleic acid sequence shown in any one of SEQ ID NO:62-68.
[0373] 6. The use according to any one of embodiments 1-5, wherein the RdCVF is human RdCVF.
[0374] 7. The use according to any one of embodiments 1-6, wherein the RdCVF comprises the amino acid sequence shown in any one of SEQ ID NO: 83-89.
...
Embodiment 1
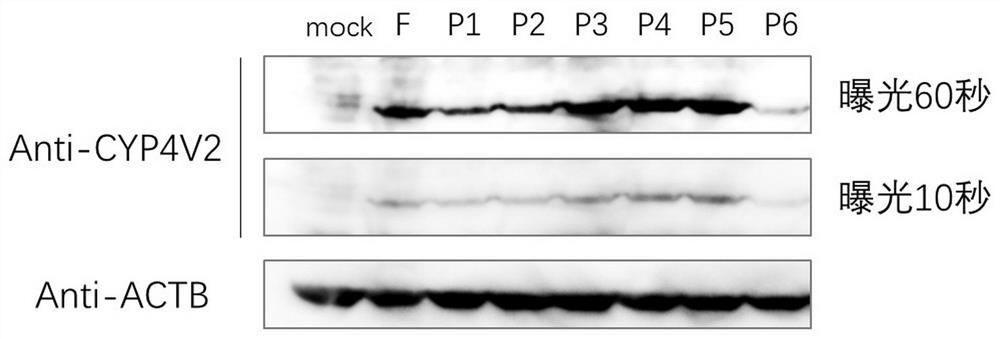
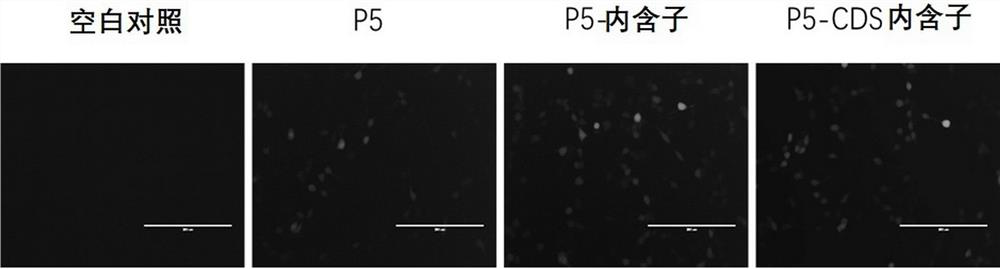
[0447] Example 1. Validation of promoter strength by expression of CYP4V2
[0448] (1) The full-length CYP4V2 promoter sequence is 2000bp upstream of the coding region of human genome CYP4V2 (HGNC:23198), synthesized by Suzhou Jinweizhi Company.
[0449] (2) Design the primer sequences of different length promoters CYP4V2-Pf, CYP4V2-P1, CYP4V2-P2, CYP4V2-P3, CYP4V2-P4, CYP4V2-P5, CYP4V2-P6, and use the nucleic acid molecules obtained in step (1) as templates PCR amplification, and gel electrophoresis to recover the amplified product. The primer sequences are shown in SEQ ID NO: 30-37.
[0450] The PCR reaction system (50 μl) is as follows:
[0451] h 2 O (Invitrogen, 10977015) 20μl
[0452] Primer F (10pmol / μl) 2μl
[0453] Primer R (10pmol / μl) 2μl
[0454] primeSTAR MAX (Takrara, R045R) 25μl
[0455] CYP4V2-pro-2000bp 5ng
[0456] Table 2 PCR reaction conditions.
[0457]
[0458] (3) The pAV-CAG-CYP4V2-P2A-EGFP vector (purchased from Shandong Weizhen Biotechnolog...
Embodiment 2
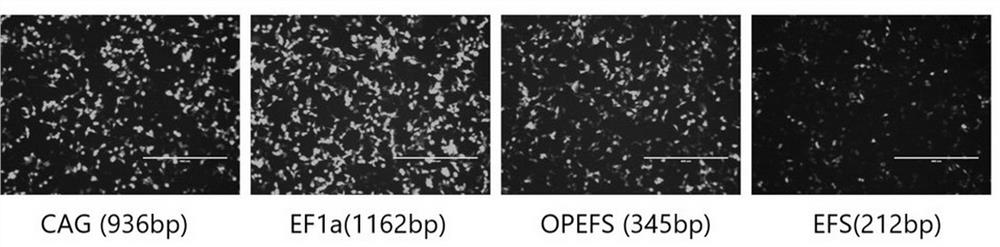
[0477] Example 2. Using EGFP as a reporter gene to verify promoter strength
[0478] (1) PCR amplification of promoters EF1a, EFS, OPEFS. The EF1a primer sequence is shown in SEQ ID NO: 40-41, and the EFS primer sequence is shown in SEQ ID NO: 42-43. The OPEFS promoter was based on the universal promoter EFS, and primers were designed to transform EFS into OPEFS by two rounds of PCR. The primers used in the first round of PCR reactions are EFS forward primers (as shown in SEQ ID NO: 42) and OPEFS-overlapR1 (as shown in SEQ ID NO: 44), and the primers used in the second round of PCR reactions are EFS forward primers ( shown in SEQ ID NO: 42) and OPEFS-overlapR2 (shown in SEQ ID NO: 45). Amplified products were recovered by gel electrophoresis. The PCR reaction system (50 μl) is as follows:
[0479] H2O (Invitrogen, 10977015) 20μl
[0480] Primer F (10pmol / μl) 2μl
[0481] Primer R (10pmol / μl) 2μl
[0482] primeSTAR MAX (Takrara, R045R) 25μl
[0483] pAV-CAG-CYP4V2-P2A-E...
PUM
| Property | Measurement | Unit |
|---|---|---|
| thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com