A yeast phenotypic screening method for isolation of functional antibodies against g-protein coupled receptors
A functional antibody and antibody technology, applied in library screening, immunoglobulin, chemical instruments and methods, etc., can solve not-so-easy problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0127] Brief overview of the platform
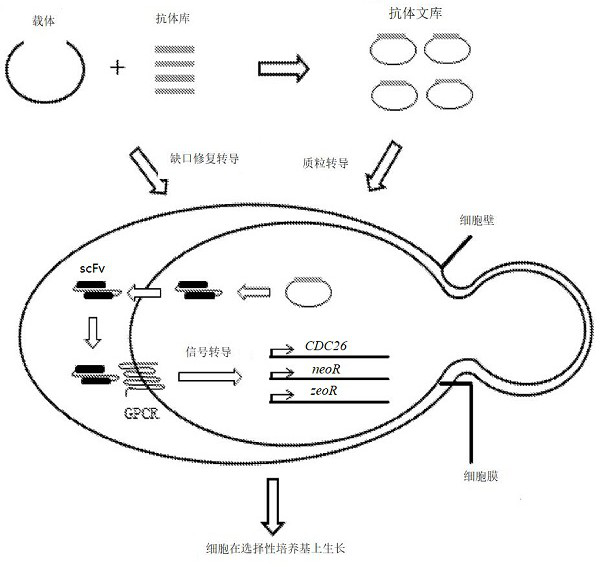
[0128] figure 1 The platform shown is typical of autocrine signaling-based phenotypic screens. Antibody libraries are introduced into cells by efficient transformation and are secreted into the periplasmic space between the cell wall and membrane. Most antibodies are retained in the periplasmic space due to their size. When the activator antibody encounters its epitope on the receptor (Ste2p or human GPCR receptor), the antibody will bind to the receptor and activate the receptor, triggering a series of kinase reactions. Activates the Stel2p transcription factor at the end of a cascade of kinase reactions. Activated Stel2p binds to the promoter regions of a panel of α-factor-induced genes and activates their transcription. The promoters of two such genes, FUS2 and FIG1, were selected to drive the expression of the reporter gene CDC26 and antibiotic resistance genes (neomycin resistance gene neoR and bleomycin resistance gene zeoR), t...
example 2
[0130] Construction of Libraries, Vectors and Host Strains
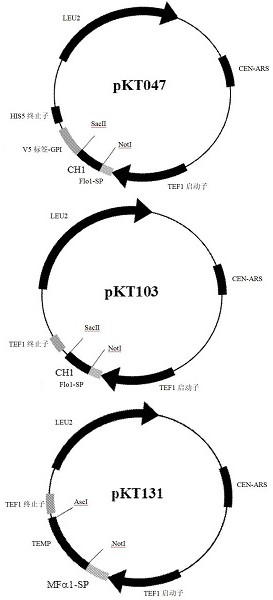
[0131] The scFv library starts with >10 10 A diversity of , which is comparable or better than most phage-displayed antibody libraries, was produced in the universal shuttle vector pRS315. Sequence analysis of randomly selected library clones indicated that more than 80% of the library members contained antibody sequences correctly assembled into the vector. When performing the screening, the library was introduced into the screening cells by PCR of the scFv library and gap repair of the vector pKT103. This process ligated the scFv fragment to the FLO1 secretion signal and placed it under the control of the TEF1 promoter.
[0132] In order to construct strains suitable for this platform, certain modifications must be made to the Ste2p signaling pathway. First, as in other experiments using the yeast α-factor signaling pathway for heterologous GPCR studies, two genes, far1Δ and sst2Δ, were knocked out to preserve c...
example 3
[0135] Ste2p activating antibody
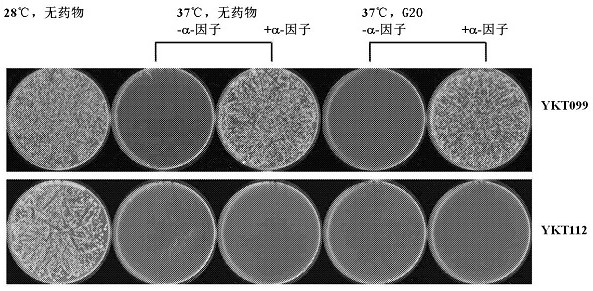
[0136] This platform was first used to find antibodies that target and activate the yeast GPCR receptor Ste2p. Use the scFv library to transform YKT099, spread the transformed cells on the SC-Leu plate (for selecting plasmids), and the SC-Leu plate also contains 20-40 μg / ml or 300-400 μg / ml of G418, respectively placed at 37 Cultivate at 30°C or 30°C. The "high temperature / low drug" selection condition was biased towards testing the expression of CDC26, while the "low temperature / high drug" condition was biased towards testing the expression of neoR. Colonies grown on the plates under each selection scheme were pooled and pooled, with each pool containing around 10 individual colonies. Plasmids were extracted from pooled cells and used again to transform YKT099. This time, transformants were screened using a reverse selection protocol. That is, if the first round of screening used high temperature / low drug selection, the second round of s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com