Method for analyzing microbial community structure based on reverse transcription of full-length small subunit ribosomal RNA
A microbial community and reverse transcription primer technology, applied in the field of biological analysis, can solve the problems of inaccurate classification of SSUrRNA gene sequences, inability to distinguish microorganisms from dead microorganisms, and unfixed sequence start sites, so as to improve bias and coverage issues, reducing experimental steps, and reducing experimental costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
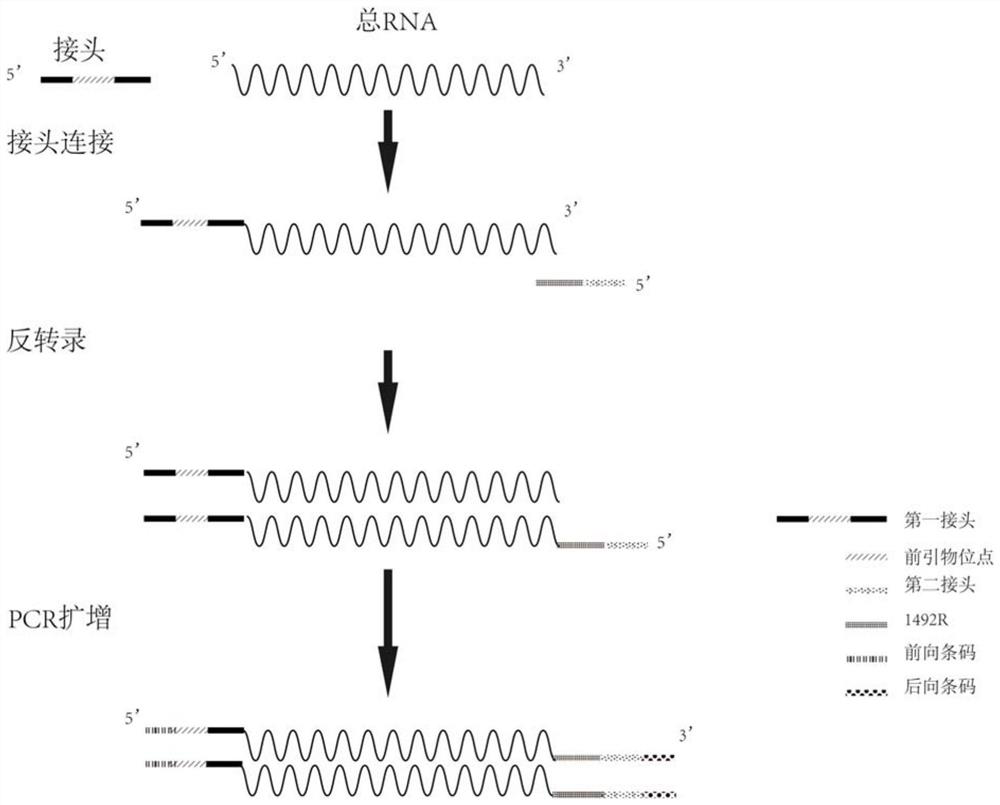
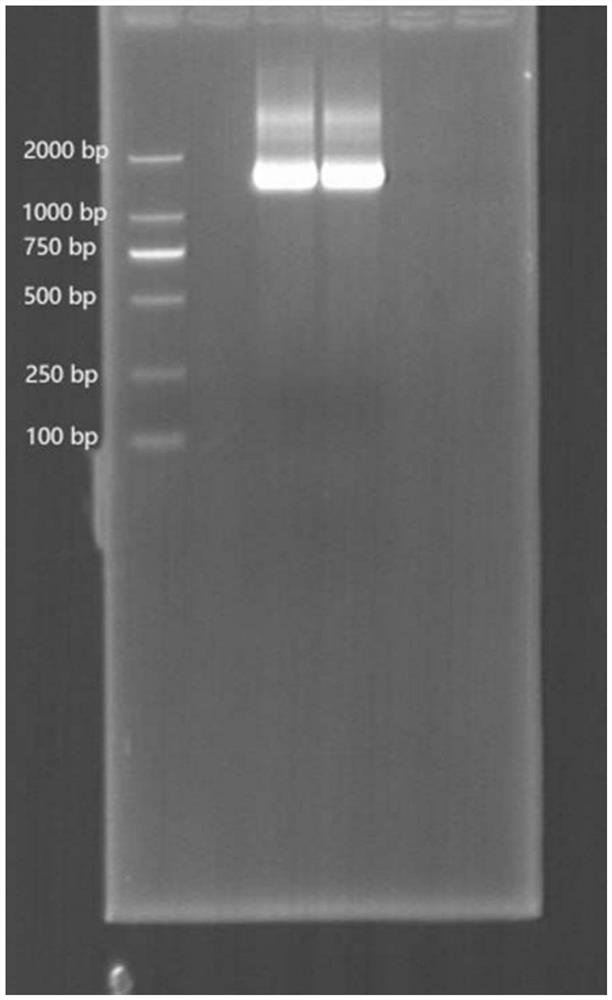
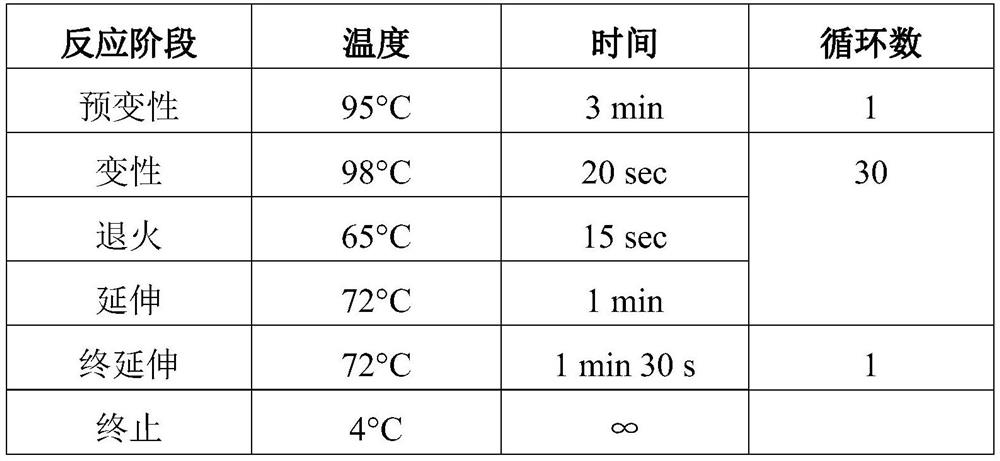
[0037] This example provides a method for analyzing microbial community structure based on reverse transcription of full-length small subunit ribosomal RNA. After extracting total RNA from environmental samples, add the first linker to the 5' end of the total RNA, and then use The reverse "universal" primer 1492R (5' end plus a second adapter) covering prokaryotic and eukaryotic SSU rRNA reverses SSU rRNA, and then uses primers designed based on the adapter sequences at both ends to perform PCR amplification to obtain ribosomal SSU cDNA, Finally, combined with third-generation sequencing, the full-length SSUrRNA sequence information was measured to obtain an accurate microbial community structure.
[0038] The specific operation process of this embodiment is as follows, as figure 1 Shown:
[0039] 1. Design reverse transcription primers, add a linker sequence 5'-CAAGCAGAAGACGGCATACGAG-3' at the 5' end of the "universal" primer 1492R sequence (5'-TACCTTGTTAYGACTT-3'), and add ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com