Acetylation of human parkin protein and application of parkin protein in medicine preparation
An acetylation and deacetylase technology, applied in the field of physiology and molecular biology, can solve undisclosed, enhanced mitophagy and anti-tumor functions, undisclosed parkin and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
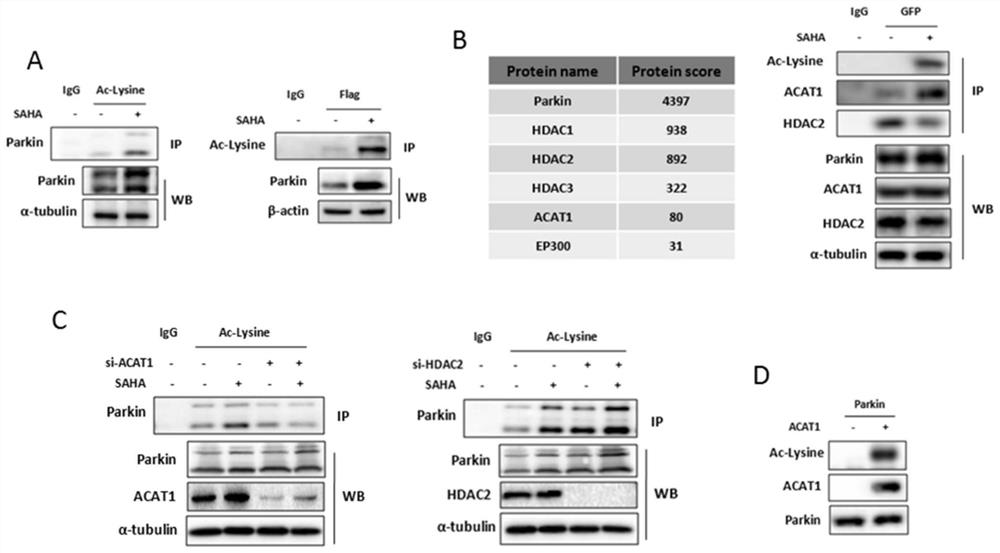
[0025] Since the acetylation of parkin protein has not been clearly reported, there is no commercial acetylation antibody for parkin. HEK293 cells were used to overexpress Flag-parkin, treated with histone deacetylase inhibitor (HDACI) SAHA drug at 2.5 μM for 12 hours, and protein samples were precipitated by FLAG or acetyl-lysine (Ac-lysine) affinity purification gel, Western blot (abbreviated as WB) detected acetyl-lysine or parkin protein levels, and found that the acetylation level of parkin in cells treated with SAHA drug was significantly increased, as shown in figure 1 As shown in A.
[0026] In order to reveal the molecular mechanism that regulates the acetylation of parkin protein, protein spectrum analysis was performed on the protein complex after GFP affinity purification gel precipitation, and the histone acetylase HAT (ACAT1, EP300) and Histone deacetylase HDAC (HDAC1, HDAC2, HDAC3), the obtained HAT and HDAC protein name and protein score (protein score), such ...
Embodiment 2
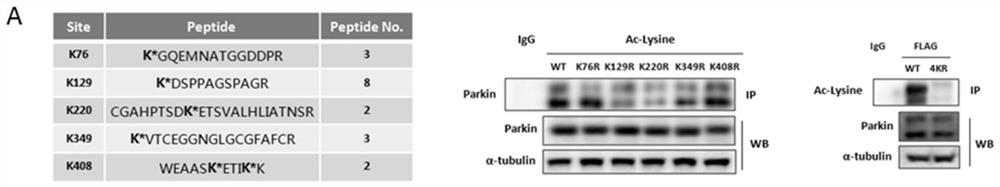
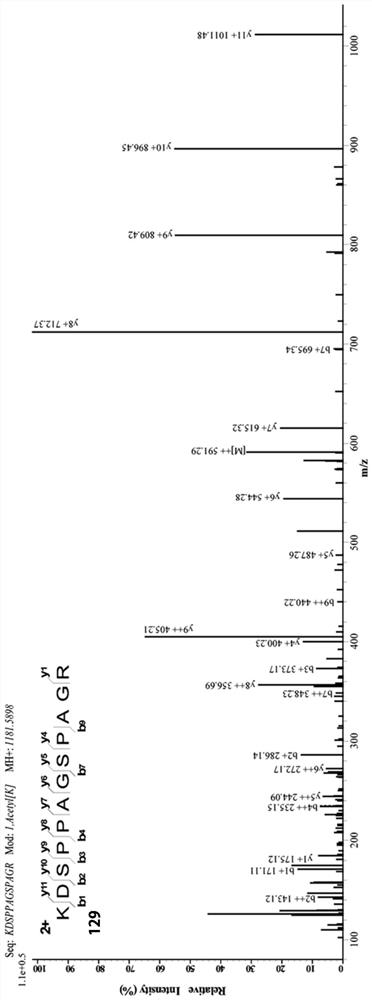
[0031] Parkin protein is mainly regulated by phosphorylation modification, and acetylation modification has not been reported. In order to further clarify the molecular mechanism regulating parkin acetylation, HEK293 cells overexpressing Flag-parkin were treated with 2.5 μM SAHA drug for 12 hours, protein samples were analyzed by SDS-PAGE gel after FLAG affinity purification gel precipitation, and parkin strips were cut out After digestion of strips and strips, the acetylation sites of parkin were found by protein spectrum LC-MS analysis. It was found that SAHA treatment caused acetylation of parkin protein sites K76, K129, K220, K349, and K408, such as Figure 2B1 , Figure 2B2 , Figure 2B3 with Figure 2B4 shown. At the same time, multiple acetylation sites screened by protein spectrum were verified and analyzed, and KR (lysine to arginine) point mutation was carried out to construct KR mutant plasmid. After HEK293 cells were transfected with parkin wild-type (parkin-WT...
Embodiment 3
[0033] Recent studies suggest that the level of mitochondrial protein acetylation is closely related to mitophagy and tumorigenesis. In order to clarify the effect of parkin protein acetylation on the level of parkin-mediated mitophagy, HeLa tumor cells were first transfected with parkin-WT plasmid and parkin-KR plasmid, and after 24 hours, the transfected HeLa tumor cells were treated with SAHA drug 2.5 μM was treated for 12 hours, and Western blot was used to detect the changes in the expression of mitochondrial membrane proteins before and after SAHA treatment, and the results were as follows image 3 As shown in A and 3B, before SAHA treatment, overexpression of parkin-WT reduces the levels of mitochondrial membrane proteins such as VDAC, MFN2, TIM23 and HSP60, and increases the levels of autophagy proteins such as LC3 and P62 in mitochondria, while the parkin acetylation site mutation After that, it weakened the degradation effect of parkin-WT on mitochondrial membrane pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com